



Biodiv Sci ›› 2021, Vol. 29 ›› Issue (11): 1505-1512. DOI: 10.17520/biods.2021166 cstr: 32101.14.biods.2021166
• Original Papers: Animal Diversity • Previous Articles Next Articles
Denggao Xiang1,2, Yuefei Li2, Xinhui Li2, Weitao Chen2,*( ), Xiuhui Ma1,*(
), Xiuhui Ma1,*( )
)
Received:2021-04-29
Accepted:2021-07-05
Online:2021-11-20
Published:2021-07-27
Contact:
Weitao Chen,Xiuhui Ma
Denggao Xiang, Yuefei Li, Xinhui Li, Weitao Chen, Xiuhui Ma. Population structure and genetic diversity of Culter recurviceps revealed by multi-loci[J]. Biodiv Sci, 2021, 29(11): 1505-1512.
| 群体 Population | 样本量 Sample size | 经纬度 Locality |
|---|---|---|
| 海南岛水系 Hainan Island drainage | ||
| 白沙 Baisha (BS) | 33 | 19˚13′ N/109˚26′ E |
| 南丰 Nanfeng (NF) | 15 | 19˚25′ N/109˚33′ E |
| 定安 Dingan (DA) | 4 | 19˚40′ N/110˚21′ E |
| 珠江水系 Pearl River drainage | ||
| 从江 Congjiang (CJ) | 9 | 25˚44′ N/108˚55′ E |
| 崇左 Chongzuo (CZ) | 14 | 22˚16′ N/107˚22′ E |
| 桂平 Guiping (GP) | 14 | 23˚23′ N/110˚04′ E |
| 武宣 Wuxuan (WX) | 2 | 23˚35′ N/109˚40′ E |
| 古竹 Guzhu (GZ) | 16 | 23˚30′ N/114˚42′ E |
| 惠州 Huizhou (HZ) | 11 | 23˚06′ N/114˚24′ E |
| 江门 Jiangmen (JM) | 4 | 22˚34′ N/113˚07′ E |
| 横沥 Hengli (HL) | 1 | 22˚43′ N/113˚29′ E |
| 开平 Kaiping (KP) | 5 | 22˚22′ N/112˚41′ E |
| 柳州 Liuzhou (LZ) | 5 | 24˚18′ N/109˚23′ E |
| 柳城 Liucheng (LC) | 8 | 24˚38′ N/109˚14′ E |
| 南宁 Nanning (NN) | 7 | 22˚46′ N/108˚21′ E |
| 清远 Qingyuan (QY) | 5 | 23˚37′ N/113˚03′ E |
| 梧州 Wuzhou (WZ) | 13 | 23˚25′ N/111˚16′ E |
| 濛江 Mengjiang (MJ) | 1 | 23˚27′ N/110˚44′ E |
| 永福 Yongfu (YF) | 12 | 24˚59′ N/109˚59′ E |
| 郁南 Yunan (YN) | 11 | 23˚14′ N/111˚31′ E |
| 思林 Silin (SL) | 12 | 23˚30′ N/107˚20′ E |
| 韶关 Shaoguan (SG) | 2 | 24˚47′ N/113˚35′ E |
| 平乐 Pingle (PL) | 3 | 24˚37′ N/110˚38′ E |
| 总计 Total | 207 | |
Table 1 Sampling information of Culter recurviceps specimens
| 群体 Population | 样本量 Sample size | 经纬度 Locality |
|---|---|---|
| 海南岛水系 Hainan Island drainage | ||
| 白沙 Baisha (BS) | 33 | 19˚13′ N/109˚26′ E |
| 南丰 Nanfeng (NF) | 15 | 19˚25′ N/109˚33′ E |
| 定安 Dingan (DA) | 4 | 19˚40′ N/110˚21′ E |
| 珠江水系 Pearl River drainage | ||
| 从江 Congjiang (CJ) | 9 | 25˚44′ N/108˚55′ E |
| 崇左 Chongzuo (CZ) | 14 | 22˚16′ N/107˚22′ E |
| 桂平 Guiping (GP) | 14 | 23˚23′ N/110˚04′ E |
| 武宣 Wuxuan (WX) | 2 | 23˚35′ N/109˚40′ E |
| 古竹 Guzhu (GZ) | 16 | 23˚30′ N/114˚42′ E |
| 惠州 Huizhou (HZ) | 11 | 23˚06′ N/114˚24′ E |
| 江门 Jiangmen (JM) | 4 | 22˚34′ N/113˚07′ E |
| 横沥 Hengli (HL) | 1 | 22˚43′ N/113˚29′ E |
| 开平 Kaiping (KP) | 5 | 22˚22′ N/112˚41′ E |
| 柳州 Liuzhou (LZ) | 5 | 24˚18′ N/109˚23′ E |
| 柳城 Liucheng (LC) | 8 | 24˚38′ N/109˚14′ E |
| 南宁 Nanning (NN) | 7 | 22˚46′ N/108˚21′ E |
| 清远 Qingyuan (QY) | 5 | 23˚37′ N/113˚03′ E |
| 梧州 Wuzhou (WZ) | 13 | 23˚25′ N/111˚16′ E |
| 濛江 Mengjiang (MJ) | 1 | 23˚27′ N/110˚44′ E |
| 永福 Yongfu (YF) | 12 | 24˚59′ N/109˚59′ E |
| 郁南 Yunan (YN) | 11 | 23˚14′ N/111˚31′ E |
| 思林 Silin (SL) | 12 | 23˚30′ N/107˚20′ E |
| 韶关 Shaoguan (SG) | 2 | 24˚47′ N/113˚35′ E |
| 平乐 Pingle (PL) | 3 | 24˚37′ N/110˚38′ E |
| 总计 Total | 207 | |
| 群体 Population | 单倍型数 Number of haplotype | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 中性检测 Neutrality tests | |
|---|---|---|---|---|---|
| Tajima’s D | Fu’s Fs | ||||
| BS | 7 | 0.383 (± 0.107) | 0.0002 (±0.0001) | -1.75856** | -6.19981*** |
| CJ | 4 | 0.806 (± 0.089) | 0.0021 (±0.0003) | 1.29983 | 2.70828 |
| CZ | 7 | 0.879 (± 0.058) | 0.0009 (±0.0001) | 0.01278 | -1.77070 |
| GP | 10 | 0.923 (± 0.060) | 0.0011 (±0.0001) | -0.27622 | -5.21979*** |
| GZ | 10 | 0.917 (± 0.049) | 0.0013 (±0.0003) | -1.10830 | -3.75174* |
| HZ | 7 | 0.891 (± 0.074) | 0.0014 (±0.0002) | -1.41142 | -1.25974 |
| KP | 4 | 0.900 (± 0.161) | 0.0019 (±0.0008) | -0.92693 | 0.35685 |
| LZ | 3 | 0.800 (± 0.164) | 0.0026 (±0.0006) | 1.25408 | 2.74649 |
| LC | 5 | 0.893 (± 0.086) | 0.0018 (±0.0005) | -0.66317 | 0.49798 |
| NF | 2 | 0.133 (± 0.112) | 0.0001 (±0.0001) | -1.15945 | -0.64899 |
| NN | 3 | 0.762 (± 0.115) | 0.0006 (±0.0002) | 1.10686 | 0.78880 |
| QY | 5 | 1.000 (± 0.126) | 0.0031 (±0.0006) | 0.40617 | -0.71434 |
| WZ | 12 | 0.987 (± 0.035) | 0.0022 (±0.0004) | -1.15138 | -6.16071*** |
| YF | 2 | 0.485 (± 0.106) | 0.0004 (±0.0001) | 1.35640 | 2.32787 |
| YN | 5 | 0.782 (± 0.107) | 0.0011 (±0.0004) | -1.13653 | 0.36244 |
| SL | 9 | 0.909 (± 0.079) | 0.0020 (±0.0004) | -0.32946 | -2.27867 |
| DA | 1 | NA | NA | NA | NA |
| SG | 2 | NA | NA | NA | NA |
| PL | 1 | NA | NA | NA | NA |
| WX | 2 | NA | NA | NA | NA |
| JM | 4 | NA | NA | NA | NA |
| MJ | 1 | NA | NA | NA | NA |
| HL | 1 | NA | NA | NA | NA |
| 总计 Total | 52 | 0.912 (± 0.012) | 0.0023 (±0.0001) | -0.91788 | -24.88587*** |
Table 2 Number of haplotype, haplotype diversity, nucleotide diversity and neutrality tests based on CCN sequences. Population codes see Table 1.
| 群体 Population | 单倍型数 Number of haplotype | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity | 中性检测 Neutrality tests | |
|---|---|---|---|---|---|
| Tajima’s D | Fu’s Fs | ||||
| BS | 7 | 0.383 (± 0.107) | 0.0002 (±0.0001) | -1.75856** | -6.19981*** |
| CJ | 4 | 0.806 (± 0.089) | 0.0021 (±0.0003) | 1.29983 | 2.70828 |
| CZ | 7 | 0.879 (± 0.058) | 0.0009 (±0.0001) | 0.01278 | -1.77070 |
| GP | 10 | 0.923 (± 0.060) | 0.0011 (±0.0001) | -0.27622 | -5.21979*** |
| GZ | 10 | 0.917 (± 0.049) | 0.0013 (±0.0003) | -1.10830 | -3.75174* |
| HZ | 7 | 0.891 (± 0.074) | 0.0014 (±0.0002) | -1.41142 | -1.25974 |
| KP | 4 | 0.900 (± 0.161) | 0.0019 (±0.0008) | -0.92693 | 0.35685 |
| LZ | 3 | 0.800 (± 0.164) | 0.0026 (±0.0006) | 1.25408 | 2.74649 |
| LC | 5 | 0.893 (± 0.086) | 0.0018 (±0.0005) | -0.66317 | 0.49798 |
| NF | 2 | 0.133 (± 0.112) | 0.0001 (±0.0001) | -1.15945 | -0.64899 |
| NN | 3 | 0.762 (± 0.115) | 0.0006 (±0.0002) | 1.10686 | 0.78880 |
| QY | 5 | 1.000 (± 0.126) | 0.0031 (±0.0006) | 0.40617 | -0.71434 |
| WZ | 12 | 0.987 (± 0.035) | 0.0022 (±0.0004) | -1.15138 | -6.16071*** |
| YF | 2 | 0.485 (± 0.106) | 0.0004 (±0.0001) | 1.35640 | 2.32787 |
| YN | 5 | 0.782 (± 0.107) | 0.0011 (±0.0004) | -1.13653 | 0.36244 |
| SL | 9 | 0.909 (± 0.079) | 0.0020 (±0.0004) | -0.32946 | -2.27867 |
| DA | 1 | NA | NA | NA | NA |
| SG | 2 | NA | NA | NA | NA |
| PL | 1 | NA | NA | NA | NA |
| WX | 2 | NA | NA | NA | NA |
| JM | 4 | NA | NA | NA | NA |
| MJ | 1 | NA | NA | NA | NA |
| HL | 1 | NA | NA | NA | NA |
| 总计 Total | 52 | 0.912 (± 0.012) | 0.0023 (±0.0001) | -0.91788 | -24.88587*** |
| BS | CJ | CZ | WG | GZ | JH | NF | HZ | KP | ZC | MZ | NN | QY | SL | YF | YN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BS | 0.000 | *** | *** | *** | *** | *** | NS | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| CJ | 0.816 | 0.000 | * | * | ** | NS | *** | NS | NS | NS | NS | NS | NS | NS | * | NS |
| CZ | 0.859 | 0.160 | 0.000 | NS | *** | NS | *** | *** | NS | * | NS | NS | *** | NS | NS | NS |
| WG | 0.838 | 0.157 | 0.000 | 0.000 | ** | * | *** | ** | NS | NS | NS | NS | * | * | NS | NS |
| GZ | 0.842 | 0.260 | 0.211 | 0.111 | 0.000 | * | *** | NS | * | * | NS | *** | *** | ** | *** | *** |
| JH | 0.849 | 0.000 | 0.237 | 0.234 | 0.307 | 0.000 | *** | * | NS | NS | NS | NS | NS | NS | * | * |
| NF | 0.000 | 0.753 | 0.834 | 0.802 | 0.803 | 0.795 | 0.000 | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| HZ | 0.868 | 0.264 | 0.288 | 0.175 | 0.025 | 0.292 | 0.830 | 0.000 | NS | * | NS | *** | ** | * | *** | *** |
| KP | 0.876 | 0.000 | 0.021 | 0.021 | 0.142 | 0.000 | 0.847 | 0.150 | 0.000 | NS | NS | NS | NS | NS | NS | NS |
| ZC | 0.747 | 0.000 | 0.091 | 0.043 | 0.103 | 0.000 | 0.664 | 0.119 | 0.000 | 0.000 | NS | NS | NS | NS | * | NS |
| MZ | 0.765 | 0.050 | 0.010 | 0.000 | 0.036 | 0.058 | 0.691 | 0.057 | 0.000 | 0.000 | 0.000 | NS | * | NS | NS | NS |
| NN | 0.899 | 0.100 | 0.083 | 0.061 | 0.290 | 0.192 | 0.906 | 0.372 | 0.136 | 0.020 | 0.078 | 0.000 | * | NS | NS | NS |
| QY | 0.841 | 0.007 | 0.351 | 0.344 | 0.373 | 0.000 | 0.779 | 0.361 | 0.046 | 0.062 | 0.146 | 0.299 | 0.000 | NS | ** | * |
| SL | 0.800 | 0.000 | 0.068 | 0.085 | 0.168 | 0.000 | 0.735 | 0.185 | 0.000 | 0.003 | 0.000 | 0.105 | 0.102 | 0.000 | NS | NS |
| YF | 0.913 | 0.211 | 0.000 | 0.024 | 0.209 | 0.324 | 0.921 | 0.314 | 0.053 | 0.113 | 0.028 | 0.227 | 0.429 | 0.066 | 0.000 | NS |
| YN | 0.854 | 0.093 | 0.000 | 0.000 | 0.161 | 0.146 | 0.821 | 0.228 | 0.000 | 0.043 | 0.000 | 0.107 | 0.218 | 0.013 | 0.000 | 0.000 |
Table 3 Pairwise population FST values (below diagonal) and significance probability estimates (above diagonal). Population codes see Table 1.
| BS | CJ | CZ | WG | GZ | JH | NF | HZ | KP | ZC | MZ | NN | QY | SL | YF | YN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BS | 0.000 | *** | *** | *** | *** | *** | NS | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| CJ | 0.816 | 0.000 | * | * | ** | NS | *** | NS | NS | NS | NS | NS | NS | NS | * | NS |
| CZ | 0.859 | 0.160 | 0.000 | NS | *** | NS | *** | *** | NS | * | NS | NS | *** | NS | NS | NS |
| WG | 0.838 | 0.157 | 0.000 | 0.000 | ** | * | *** | ** | NS | NS | NS | NS | * | * | NS | NS |
| GZ | 0.842 | 0.260 | 0.211 | 0.111 | 0.000 | * | *** | NS | * | * | NS | *** | *** | ** | *** | *** |
| JH | 0.849 | 0.000 | 0.237 | 0.234 | 0.307 | 0.000 | *** | * | NS | NS | NS | NS | NS | NS | * | * |
| NF | 0.000 | 0.753 | 0.834 | 0.802 | 0.803 | 0.795 | 0.000 | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| HZ | 0.868 | 0.264 | 0.288 | 0.175 | 0.025 | 0.292 | 0.830 | 0.000 | NS | * | NS | *** | ** | * | *** | *** |
| KP | 0.876 | 0.000 | 0.021 | 0.021 | 0.142 | 0.000 | 0.847 | 0.150 | 0.000 | NS | NS | NS | NS | NS | NS | NS |
| ZC | 0.747 | 0.000 | 0.091 | 0.043 | 0.103 | 0.000 | 0.664 | 0.119 | 0.000 | 0.000 | NS | NS | NS | NS | * | NS |
| MZ | 0.765 | 0.050 | 0.010 | 0.000 | 0.036 | 0.058 | 0.691 | 0.057 | 0.000 | 0.000 | 0.000 | NS | * | NS | NS | NS |
| NN | 0.899 | 0.100 | 0.083 | 0.061 | 0.290 | 0.192 | 0.906 | 0.372 | 0.136 | 0.020 | 0.078 | 0.000 | * | NS | NS | NS |
| QY | 0.841 | 0.007 | 0.351 | 0.344 | 0.373 | 0.000 | 0.779 | 0.361 | 0.046 | 0.062 | 0.146 | 0.299 | 0.000 | NS | ** | * |
| SL | 0.800 | 0.000 | 0.068 | 0.085 | 0.168 | 0.000 | 0.735 | 0.185 | 0.000 | 0.003 | 0.000 | 0.105 | 0.102 | 0.000 | NS | NS |
| YF | 0.913 | 0.211 | 0.000 | 0.024 | 0.209 | 0.324 | 0.921 | 0.314 | 0.053 | 0.113 | 0.028 | 0.227 | 0.429 | 0.066 | 0.000 | NS |
| YN | 0.854 | 0.093 | 0.000 | 0.000 | 0.161 | 0.146 | 0.821 | 0.228 | 0.000 | 0.043 | 0.000 | 0.107 | 0.218 | 0.013 | 0.000 | 0.000 |
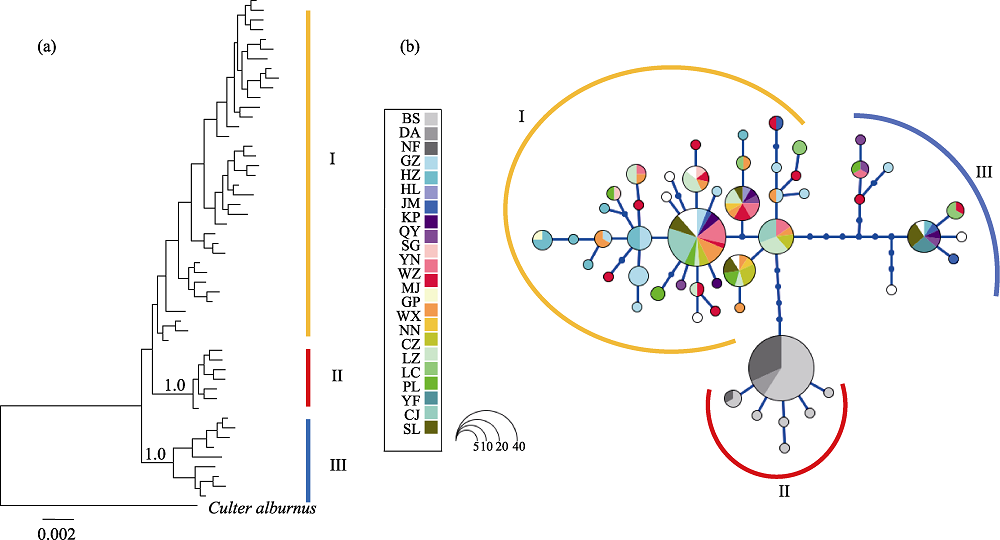
Fig. 2 Bayesian tree and haplotype network. (a) Bayesian tree for Culter recurviceps (numbers indicated posterior probability); (b) Haplotype network of C. recurviceps. Cycle size is roughly proportional to the haplotype frequency. Population codes see Table 1.
| [1] |
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC (1987) Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annual Review of Ecology and Systematics, 18, 489-522.
DOI URL |
| [2] | Chen WT, Li C, Chen FC, Li YF, Yang JP, Li J, Li XH (2020) Phylogeographic analyses of a migratory freshwater fish (Megalobrama terminalis) reveal a shallow genetic structure and pronounced effects of sea-level changes. Gene, 737, 144478. |
| [3] |
Chen XL, Chiang TY, Lin HD, Zheng HS, Shao KT, Zhang Q, Hsu KC (2007) Mitochondrial DNA phylogeography of Glyptothorax fokiensis and Glyptothorax hainanensis in Asia. Journal of Fish Biology, 70, 75-93.
DOI URL |
| [4] |
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214.
PMID |
| [5] |
Durand JD, Tsigenopoulos CS, Ünlü E, Berrebi P (2002) Erratum to “Phylogeny and Biogeography of the Family Cyprinidae in the Middle East Inferred from Cytochrome b DNA-Evolutionary Significance of This Region”. Molecular Phylogenetics and Evolution, 22, 91-100.
PMID |
| [6] |
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567.
DOI PMID |
| [7] |
Fu YX (1996) Estimating the age of the common ancestor of a DNA sample using the number of segregating sites. Genetics, 144, 829-838.
PMID |
| [8] |
Gascoyne M, Benjamin GJ, Schwarcz HP, Ford DC (1979) Sea-level lowering during the Illinoian glaciation: Evidence from a Bahama “Blue Hole”. Science, 205, 806-808.
PMID |
| [9] |
Hey J (2010) Isolation with migration models for more than two populations. Molecular Biology and Evolution, 27, 905-920.
DOI URL |
| [10] |
Ketmaier V, Bianco PG, Cobolli M, Krivokapic M, Caniglia R,De Matthaeis E (2004) Molecular phylogeny of two lineages of Leuciscinae cyprinids (Telestes and Scardinius) from the peri-Mediterranean area based on cytochrome b data. Molecular Phylogenetics and Evolution, 32, 1061-1071.
PMID |
| [11] |
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111-120.
PMID |
| [12] |
Li J, Li XH, Tan XC, Li YF, He MF, Luo JR, Lin JZ, Su SF (2009) Species diversity of fish community of Provincial Xijiang River Rare Fishes Natural Reserve in Zhaoqing City, Guangdong Province. Journal of Lake Sciences, 21, 556-562. (in Chinese with English abstract)
DOI URL |
| [ 李捷, 李新辉, 谭细畅, 李跃飞, 何美峰, 罗建仁, 林建志, 苏少芳 (2009) 广东肇庆西江珍稀鱼类省级自然保护区鱼类多样性. 湖泊科学, 21, 556-562.] | |
| [13] |
Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452.
DOI PMID |
| [14] |
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: Combining landscape ecology and population genetics. Trends in Ecology & Evolution, 18, 189-197.
DOI URL |
| [15] |
Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey J (2004) Bayesian phylogenetic analysis of combined data. Systematic Biology, 53, 47-67.
PMID |
| [16] | Qiu CF, Lin YG, Qing N, Zhao J, Chen XL (2008) Genetic variation and phylogeography of Micronoemacheilus pulcher populations among basin systems between western South China and Hainan Island. Acta Zoologica Sinica, 54, 805-813. (in Chinese with English abstract) |
| [ 丘城锋, 林岳光, 庆宁, 赵俊, 陈湘粦 (2008) 华南西部及海南岛美丽小条鳅种群遗传变异与亲缘地理. 动物学报, 54, 805-813.] | |
| [17] |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572-1574.
PMID |
| [18] | Salzburger W, Ewing GB, Von Haeseler A (2011) The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Molecular Ecology, 20, 1952-1963. |
| [19] | Shuai FM, Li XF, Liu QF, Li YF, Yang JP, Li J, Chen FC (2017) Spatial patterns of fish diversity and distribution in the Pearl River. Acta Ecologica Sinica, 37, 3182-3192. (in Chinese with English abstract) |
| [ 帅方敏, 李新辉, 刘乾甫, 李跃飞, 杨计平, 李捷, 陈方灿 (2017) 珠江水系鱼类群落多样性空间分布格局. 生态学报, 37, 3182-3192.] | |
| [20] |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595.
PMID |
| [21] |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725-2729.
DOI URL |
| [22] |
Wright S (1943) Isolation by distance. Genetics, 28, 114-138.
PMID |
| [23] | Xie G, Pang SX, Xu SY, Ye X, Qi BL, Pan DB (1998) Artificial reproduction and embryo development of Culter recurviceps. Fisheries Science and Technology, (5), 23-25. (in Chinese) |
| [ 谢刚, 庞世勋, 许淑英, 叶星, 祁宝仑, 潘德博 (1998) 海南红鲌人工繁殖和胚胎发育. 水产科技, (5), 23-25.] | |
| [24] | Xu SY, Xie G, Qi BL, Ye X, Pang SX (2001) Study on the sensitivity of Culter recurviceps to aquatic drugs. Reservoir Fisheries, 21(1), 36-36. (in Chinese) |
| [ 许淑英, 谢刚, 祁宝仑, 叶星, 庞世勋 (2001) 海南红鲌对水产药物的敏感性研究. 水利渔业, 21(1), 36-37.] | |
| [25] | Xuan ZY, Jiang T, Liu HB, Chen XB, Yang J (2020) Genetic divergence of Coilia nasus taihuensis and Coilia brachygnathus populations based on the mitochondrial Cyt-b gene. Progress in Fishery Sciences, 41(4), 33-40. (in Chinese with English abstract) |
| [ 轩中亚, 姜涛, 刘洪波, 陈修报, 杨健 (2020) 基于线粒体Cyt-b序列的太湖湖鲚与短颌鲚种群遗传分析. 渔业科学进展, 41(4), 33-40.] | |
| [26] |
Yang JQ, Hsu KC, Liu ZZ, Su LW, Kuo PH, Tang WQ, Zhou ZC, Liu D, Bao BL, Lin HD (2016) The population history of Garra orientalis (Teleostei: Cyprinidae) using mitochondrial DNA and microsatellite data with approximate Bayesian computation. BMC Evolutionary Biology, 16, 73.
DOI URL |
| [27] | Zheng CY (1989) The Ichthyography of the Pearl River. Science Press, Beijing. (in Chinese) |
| [ 郑慈英 (1989) 珠江鱼类志. 科学出版社, 北京.] | |
| [28] | Zhou ZX, Liu B, Gong J, Bai YL, Yang JY, Xu P (2020) Phylogeny and population genetics of species in Takifugu genus based on mitochondrial genome. Journal of Fisheries of China, 44, 1792-1803. (in Chinese with English abstract) |
| [ 周志雄, 刘波, 宫杰, 白玉麟, 阳俊逸, 徐鹏 (2020) 基于线粒体基因组的东方鲀属系统发育学及群体遗传学. 水产学报, 44, 1792-1803.] |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [9] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [10] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [11] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [12] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [13] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [14] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| [15] | Xinyu Cai, Xiaowei Mao, Yiqiang Zhao. Methods and research progress on the origin of animal domestication [J]. Biodiv Sci, 2022, 30(4): 21457-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn