



Biodiv Sci ›› 2015, Vol. 23 ›› Issue (6): 784-792. DOI: 10.17520/biods.2015075 cstr: 32101.14.biods.2015075
Special Issue: 青藏高原生物多样性与生态安全
• Original Papers: Animal Diversity • Previous Articles Next Articles
Lili Xie, Lei Xu*, Qiuqi Lin, Boping Han
Received:2015-03-25
Accepted:2015-09-01
Online:2015-11-20
Published:2015-12-02
Contact:
Xu Lei
Lili Xie, Lei Xu, Qiuqi Lin, Boping Han. Phylogenetics of the Daphnia longispina complex in Tibetan lakes[J]. Biodiv Sci, 2015, 23(6): 784-792.
| 采样点 Location | 纬度 经度 Latitude Longitude | 代号 Code | GenBank登录号 GenBank accession | 序列来源 Sequence source |
|---|---|---|---|---|
| 盔形溞 Daphnia galeata | ||||
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE1 | 本研究 This study | |
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE2 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC1 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC2 | 本研究 This study | |
| 骆马湖 Luoma Lake, China | 34.07º N 118.11° E | HC3 | KM555356 | GenBank |
| 西湖 West Lake, China | 30.15º N 120.08° E | HC2 | KM555355 | GenBank |
| 宝应湖 Baoying Lake, China | 33.10º N 119.14° E | HC1 | KM555354 | GenBank |
| 颈齿溞 Daphnia dentifera | ||||
| 沉错 Chencuo, China | 28.88º N 90.47° E | CC | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS1 | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS2 | 本研究 This study | |
| 那曲湖 Naqu Lake, China | 31.48º N 92.05° E | NQHA | KM555369 | GenBank |
| Canada | CND | FJ427488 | GenBank | |
| 格桑桥 Gesangqiao, China | 29.65º N 91.12° E | GSQA | KM555366 | GenBank |
| 长刺溞 Daphnia longispina | ||||
| 班公错 Bangongcuo, China 班公错 Bangongcuo, China | 33.71º N 78.81° E 33.71º N 78.81° E | BGC1 BGC2 | 本研究 This study 本研究 This study | |
| Sweden Germany Switzerland | DEM H29 SZL | EF375861 EF375860 EF375862 | GenBank GenBank GenBank |
Table 1 Geographic and genetic characteristics of 18 Daphnia longispina complex populations
| 采样点 Location | 纬度 经度 Latitude Longitude | 代号 Code | GenBank登录号 GenBank accession | 序列来源 Sequence source |
|---|---|---|---|---|
| 盔形溞 Daphnia galeata | ||||
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE1 | 本研究 This study | |
| 错鄂 Cuoe, China | 31.63º N 88.67° E | CE2 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC1 | 本研究 This study | |
| 打加芒错 Dajiamangcuo, China | 29.65º N 85.75° E | DJMC2 | 本研究 This study | |
| 骆马湖 Luoma Lake, China | 34.07º N 118.11° E | HC3 | KM555356 | GenBank |
| 西湖 West Lake, China | 30.15º N 120.08° E | HC2 | KM555355 | GenBank |
| 宝应湖 Baoying Lake, China | 33.10º N 119.14° E | HC1 | KM555354 | GenBank |
| 颈齿溞 Daphnia dentifera | ||||
| 沉错 Chencuo, China | 28.88º N 90.47° E | CC | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS1 | 本研究 This study | |
| 拉萨河 Lasahe, China | 29.68º N 91.28° E | LS2 | 本研究 This study | |
| 那曲湖 Naqu Lake, China | 31.48º N 92.05° E | NQHA | KM555369 | GenBank |
| Canada | CND | FJ427488 | GenBank | |
| 格桑桥 Gesangqiao, China | 29.65º N 91.12° E | GSQA | KM555366 | GenBank |
| 长刺溞 Daphnia longispina | ||||
| 班公错 Bangongcuo, China 班公错 Bangongcuo, China | 33.71º N 78.81° E 33.71º N 78.81° E | BGC1 BGC2 | 本研究 This study 本研究 This study | |
| Sweden Germany Switzerland | DEM H29 SZL | EF375861 EF375860 EF375862 | GenBank GenBank GenBank |
| HC3 | HC1 | HC2 | CE | DJMC | |
|---|---|---|---|---|---|
| HC3 | n.c. | ||||
| HC1 | 0.33 ± 0.23 | n.c. | |||
| HC2 | 2.15 ± 0.57 | 1.81 ± 0.53 | n.c. | ||
| CE | 2.74 ± 0.66 | 2.40 ± 0.63 | 0.90 ± 0.36 | 0.16 ± 0.15 | |
| DJMC | 2.49 ± 0.62 | 2.15 ± 0.59 | 0.65 ± 0.33 | 0.90 ± 0.37 | 0.16 ±0.15 |
Table 2 The genetic differentiation (%) between (below the diagonal) and within (on the diagonal) Daphnia galeata populations
| HC3 | HC1 | HC2 | CE | DJMC | |
|---|---|---|---|---|---|
| HC3 | n.c. | ||||
| HC1 | 0.33 ± 0.23 | n.c. | |||
| HC2 | 2.15 ± 0.57 | 1.81 ± 0.53 | n.c. | ||
| CE | 2.74 ± 0.66 | 2.40 ± 0.63 | 0.90 ± 0.36 | 0.16 ± 0.15 | |
| DJMC | 2.49 ± 0.62 | 2.15 ± 0.59 | 0.65 ± 0.33 | 0.90 ± 0.37 | 0.16 ±0.15 |
| DEM | H29 | SZL | BGC | |
|---|---|---|---|---|
| DEM | n.c. | |||
| H29 | 1.81 ± 0.53 | n.c. | ||
| SZL | 1.81 ± 0.54 | 1.31 ± 0.43 | n.c. | |
| BGC | 5.50 ± 0.92 | 4.88 ± 0.84 | 4.62 ± 0.80 | 1.62 ± 0.51 |
Table 3 The genetic differentiation (%) between (below the diagonal) and within (on the diagonal) Daphnia longispina populations
| DEM | H29 | SZL | BGC | |
|---|---|---|---|---|
| DEM | n.c. | |||
| H29 | 1.81 ± 0.53 | n.c. | ||
| SZL | 1.81 ± 0.54 | 1.31 ± 0.43 | n.c. | |
| BGC | 5.50 ± 0.92 | 4.88 ± 0.84 | 4.62 ± 0.80 | 1.62 ± 0.51 |
| CND | NQHA | GSQA | LS | CC | |
|---|---|---|---|---|---|
| CND | n.c. | ||||
| NQHA | 1.98 ± 0.55 | n.c. | |||
| GSQA | 2.15 ± 0.60 | 0.49 ± 0.30 | n.c. | ||
| LS | 2.32 ± 0.60 | 0.65 ± 0.30 | 0.33 ± 0.16 | 0.65 ± 0.32 | |
| CC | 2.15 ± 0.60 | 0.49 ± 0.30 | 0.00 | 0.33 ± 0.16 | n.c. |
Table 4 The genetic differentiation (%) between (below the diagonal) and within (on the diagonal) Daphnia dentifera populations
| CND | NQHA | GSQA | LS | CC | |
|---|---|---|---|---|---|
| CND | n.c. | ||||
| NQHA | 1.98 ± 0.55 | n.c. | |||
| GSQA | 2.15 ± 0.60 | 0.49 ± 0.30 | n.c. | ||
| LS | 2.32 ± 0.60 | 0.65 ± 0.30 | 0.33 ± 0.16 | 0.65 ± 0.32 | |
| CC | 2.15 ± 0.60 | 0.49 ± 0.30 | 0.00 | 0.33 ± 0.16 | n.c. |
| DG | DL | DD | |
|---|---|---|---|
| DG | 1.46 ± 0.31 | 71(67.62%) | 79(80.61%) |
| DL | 16.70 ± 1.72 | 3.66 ± 0.59 | 32(49.23%) |
| DD | 16.98 ± 1.87 | 9.40 ± 1.12 | 1.01 ± 0.24 |
Table 5 The genetic differentiation (%) between and within species in Daphnia longispina complex (on the diagonal and below the diagonal) and the fixed differences among these haplotypes (above the diagonal)
| DG | DL | DD | |
|---|---|---|---|
| DG | 1.46 ± 0.31 | 71(67.62%) | 79(80.61%) |
| DL | 16.70 ± 1.72 | 3.66 ± 0.59 | 32(49.23%) |
| DD | 16.98 ± 1.87 | 9.40 ± 1.12 | 1.01 ± 0.24 |
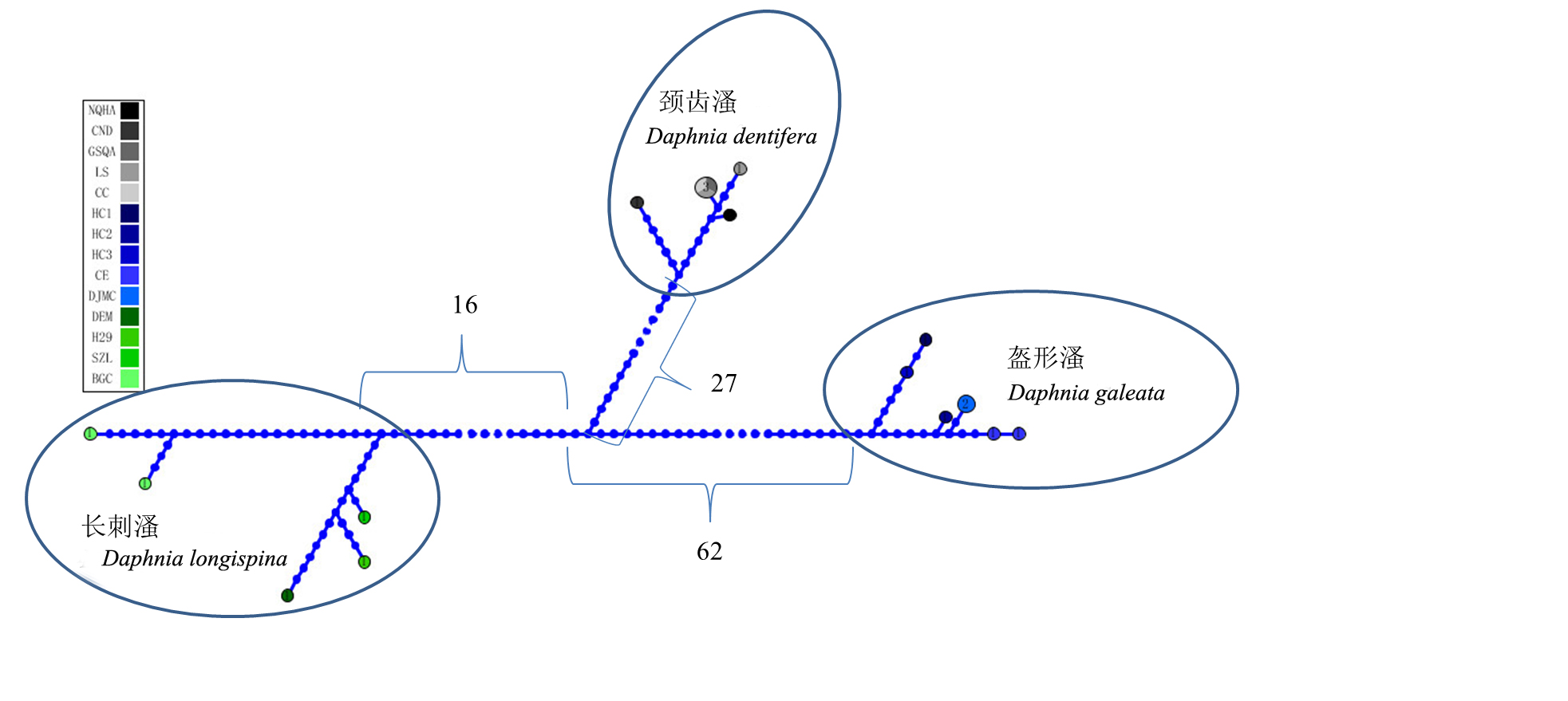
Fig. 1 Haplotype network for Daphnia longispina complex based on mitochondria COI gene sequences. Numbers between subnetworks represent the average number of mutations between subclades. The population codes are the same as those in Table 1.
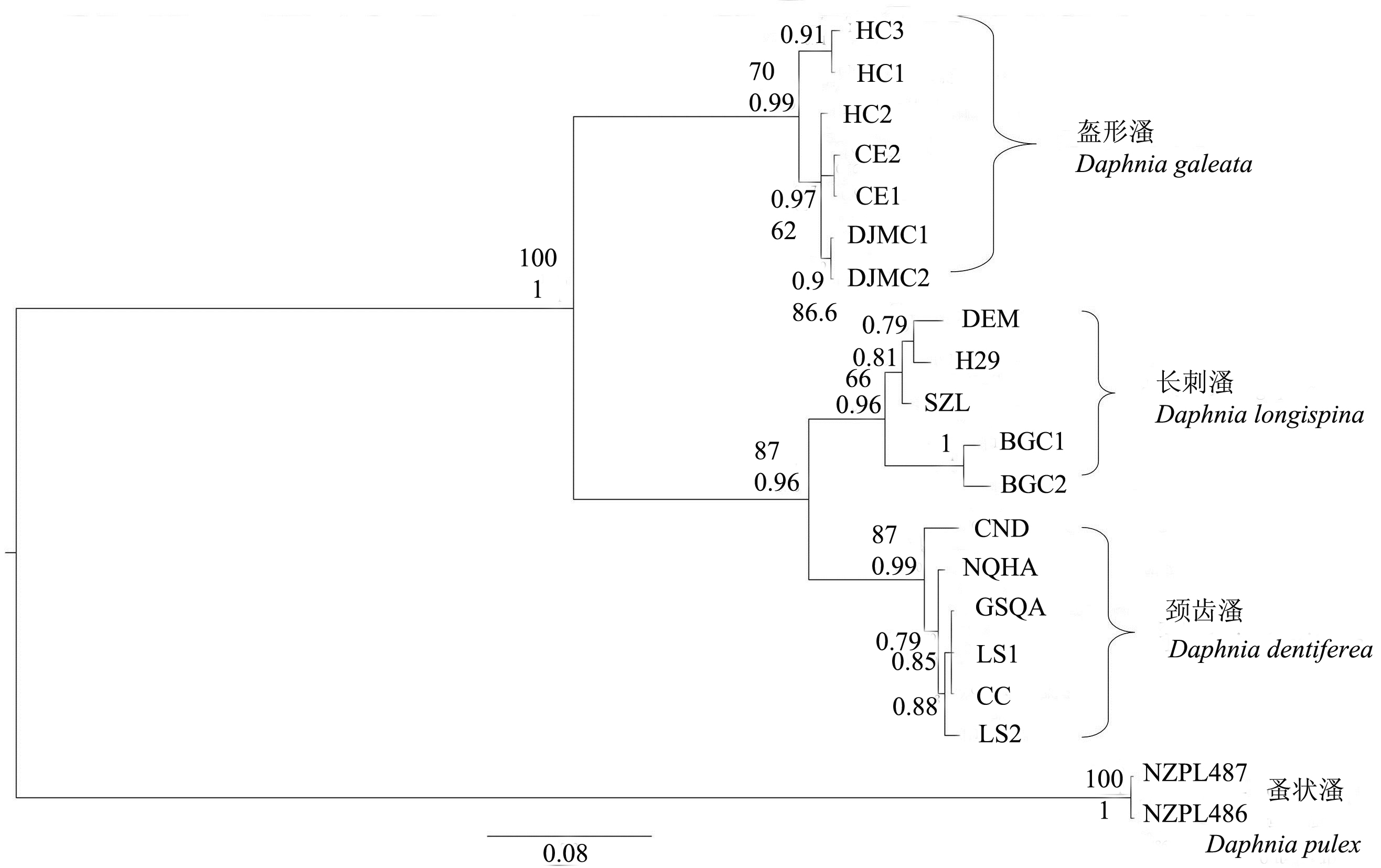
Fig. 2 Bayes and Maximum Likelihood phylogenetic tree of Daphnia longispina complex based on mitochondria COI gene sequences. Posterior probabilities (Bayes tree) and bootstrap values (Maximum Likelihood tree) are shown above the nodes. The population codes are the same as those in Table 1.
| 1 | Adamowicz SJ, Purvis A (2005) How many branchiopod crustacean species are there? Quantifying the components of underestimation.Global Ecology and Biogeography, 14, 455-468. |
| 2 | Adamowicz SJ, Petrusek A, Colbourne JK, Hebert PDN, Witt JDS (2009) The scale of divergence: a phylogenetic appraisal of intercontinental allopatric speciation in a passively dispersed freshwater zooplankton genus.Molecular Phylogenetics and Evolution, 50, 423-436. |
| 3 | Allen MR, Thum RA, Caceres CE (2010) Does local adaptation to resources explain genetic differentiation among Daphnia populations?Molecular Ecology, 19, 3076-3087. |
| 4 | Benzie JAH (1988) The systematics of Australian Daphnia (Cladocera: Daphniidae). Electrophoretic analyses of the Daphnia carinata complex.Hydrobiologia, 166, 183-197. |
| 5 | Benzie JAH (2005) Cladocera: the genus Daphnia (including Daphniopsis) (Anomopoda: Daphniidae). Guides to the identification of the microinvertebrates of the continental waters of the world.Quarterly Review of Biology, 80, 491-510. |
| 6 | Chiang SC (蒋燮治), Du NS (堵南山) (1979) Fauna Sinica, Crustacea, Freshwater Cladocera (中国动物志·淡水枝角类). Science Press, Beijing. (in Chinese) |
| 7 | Chiang SC (蒋燮治), Shen YF (沈韫芬), Gong XJ (龚循矩) (1983) Aquatic Invertebrates of the Tibetan Plateau (西藏水生无脊椎动物). Science Press, Beijing. (in Chinese) |
| 8 | Colbourne JK, Crease TJ, Weider LJ, Hebert PDN, Dufresne F, Hobaek A (1998) Phylogenetics and evolution of a circumarctic species complex (Cladocera: Daphnia pulex).Biological Journal of the Linnean Society, 65, 347-365. |
| 9 | Costa FO, Jeremy RD, James B, Rantnasingham S, Dooh RT, Hajibaei M, Hebert PDN (2007) Biological identifications through DNA barcodes: the case of the Crustacea.Canadian Journal of Fisheries and Aquatic Sciences, 64, 272-290. |
| 10 | De Meester L (1996) Local genetic differentiation and adaptation in freshwater zooplankton populations: patterns and processes.Ecoscience, 3, 385-399. |
| 11 | Dlouhá S, Thielsch A, Kraus RHS, Seda J, Schwenk K, Petrusek A (2010) Identifying hybridizing taxa within the Daphnia longispina species complex: which methods to rely on?Hydrobiologia, 643, 107-122. |
| 12 | Dodson S (1990) Predicting diel vertical migration of zooplankton.Limnology and Oceanography, 35, 1195-1200. |
| 13 | Dumont HJ, Negrea S (2002) Introduction to the Class Brachiopoda, Guides to the Identification of the Microinvertebrates of the Continental Waters of World 19. Backhuys Publishers, Leiden. |
| 14 | Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates.Molecular Marine Biology and Biotechnology, 3, 294-299. |
| 15 | Forro′ L, Korovchinsky NM, Kotov AA, Petrusek A (2008) Global diversity of cladocerans (Cladocera; Crustacea) in freshwater. Hydrobiologia, 595, 177-184. |
| 16 | Giessler S (1997a) Analysis of reticulate relationships within the Daphnia longispina species complex. Allozyme phenotype and morphology.Journal of Evolutionary Biology, 10, 87-105. |
| 17 | Giessler S (1997b) Gene flow in the Daphnia longispina hybrid complex (Crustacea, Cladocera) inhabiting large lakes.Heredity, 79, 231-241. |
| 18 | Hebert PHD (1978) The population biology of Daphnia (Crustacea, Daphnidae).Biological Reviews, 53, 387-426. |
| 19 | Hebert PDN, Cywinska A, Ball SL, De Waard JR (2003a) Biological identifications through DNA barcodes.Proceedings of the Royal Society B: Biological Sciences, 270, 313-321. |
| 20 | Hebert PDN, Witt JDS, Adamowicz SJ (2003b) Phylogeographical patterning in Daphnia ambigua: regional divergence and intercontinental cohesion.Limnology and Oceanography, 48, 261-268. |
| 21 | Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogeny.Bioinformatics, 17, 754-755. |
| 22 | Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science, 294, 2310-2314. |
| 23 | Hutchison GE (1967) A Treatise on Limnology. PhD dissertation, National Academy of Sciences, New York. |
| 24 | Irestedt M, Fjeldså J, Nylander J, Ericson P (2004) Phylogenetic relationships of typical antbirds (Thamnophilidae) and test of incongruence based on Bayes factors.BMC Evolutionary Biology, 4, 1-16. |
| 25 | Ishida S, Taylor DJ (2007) Quaternary diversification in a sexual Holarctic zooplankter, Daphnia galeata.Molecular Ecology, 16, 569-582. |
| 26 | Ishida S, Takahashi A, Matsushima N, Yokoyama J, Makino W, Urabe J, Kawata M (2011) The long-term consequences of hybridization between the two Daphnia species, D. galeata and D. dentifera, in mature habitats.BMC Evolutionary Biology, 11, 209-222. |
| 27 | Korovchinsky NM (1996) How many species of Cladocera are there?Hydrobiologia, 321, 191-204. |
| 28 | Kotov AA (2015) A critical review of the current taxonomy of the genus Daphnia O. F. Müller, 1785 (Anomopoda, Cladocera).Zootaxa, 3911, 184-200. |
| 29 | Kotov AA, Ishida S, Taylor DJ (2006) A new species in the Daphnia curvirostris (Crustacea: Cladocera) complex from the eastern Palearctic with molecular phylogenetic evidence for the independent origin of neckteeth.Journal of Plankton Research, 28, 1067-1079. |
| 30 | Kumar S, Dudley J, Nei M, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequence.Briefings in Bioinformatics, 9, 299-306. |
| 31 | Lampert W (2011) Daphnia: Development of Model Organism in Ecology and Evolution. American Institute of Biological Sciences, Washington DC. |
| 32 | Ma XL, Petrusek A, Wolinska J, Gießler S, Zhong Y, Hu W, Yin MB (2015) Diversity of the Daphnia longispina species complex in Chinese lakes: a DNA taxonomy approach.Journal of Plankton Research, 37, 56-65. |
| 33 | Mergeay J, Aguilera X, Declerck S, Petrusek A, Huyse T, De Meester L (2008) The genetic legacy of polyploid Bolivian Daphnia: the tropical Andes as a source for the North and South American Daphnia pulicaria complex.Molecular Ecology, 17, 1789-1800. |
| 34 | Mőst M, Petrusek A, Sommaruga R, Juračka PJ, Slusarczyk M, Manca M (2013) At the edge and on the top: molecular identification and ecology of Daphnia dentifera and D. longispina in high-altitude Asian lakes.Hydrobiologia, 715, 169-180. |
| 35 | Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers.Computer Applications in the Biosciences, 12, 357-358. |
| 36 | Petrusek A, Cerny M, Mergeay J, Schwenk K (2007) Daphnia in the Tatra Mountain lakes: multiple colonization and hidden species diversity revealed by molecular markers.Fundamental and Applied Limnology, 169, 279-291. |
| 37 | Petrusek A, Hobæk A, Nilssen JP, Skage M, Černý M, Brede N, Schwenk K (2008) A taxonomic reappraisal of the European Daphnia longispina complex (Crustacea, Cladocera, Anomopoda).Zoologica Scripta, 37, 507-519. |
| 38 | Petrusek A (2007) Diversity of European Daphnia on Different Scales: from Cryptic Species to Within-lake Differentiation. PhD dissertation, Charles University, Prague. |
| 39 | Posada D, Crandall KA (2001) Selecting the best-fit model of nucleotide substitution.Systematic Biology, 50, 580-601. |
| 40 | Rozas J, Sanchez-Delbarria JC, Messeguer X (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods.Bioinformatics, 19, 2496-2497. |
| 41 | Salzburger W, Ewing GB, Haeseler VA (2011) The performance of phylogenetic algorithms in estimating haplotype genealogies with migration.Molecular Ecology, 20, 1952-1963. |
| 42 | Sekino T, Yoshioka T (1995) The relationship between nutritional condition and diel vertical migration of Daphnia galeata.Japanese Journal of Limnology, 56, 145-150. |
| 43 | Skage M, Hobæk A, Ruthová Š, Keller B, Petrusek A, Sed’a J, Spaak P (2007) Intra-specific rDNA-ITS restriction site variation and an improved protocol to distinguish species and hybrids in the Daphnia longispina complex.Hydrobiologia, 594, 19-32. |
| 44 | Sommer RS, Zachos FE (2009) Fossil evidence and phylogeography of temperate species: glacial refugia and post-glacial recolonization.Journal of Biogeography, 36, 2013-2020. |
| 45 | Spaak P, Fox J, Hairston NG Jr (2012) Modes and mechanisms of a Daphnia invasion.Proceedings of the Royal Society B: Biological Sciences, 279, 2936-2944. |
| 46 | Swofford D (2003) PAUP* 4.0 |
| [Electronic Resource]: Phylogenetic Analysis Using Parsimony. Sinauer Associates Publishers, Sunderland. | |
| 47 | Taylor DJ, Hebert PD, Colbourne JK (1996) Phylogenetics and evolution of the Daphnia longispina group (Crustacea) based on 12S rDNA sequence and allozyme variation.Molecular Phylogenetics and Evolution, 5, 495-510. |
| 48 | Taylor DJ, Finston TL, Hebert PDN (1998) Biogeography of a widespread freshwater crustacean: pseudocongruence and cryptic endemism in the North American Daphnia laevis complex.Evolution, 52, 1648-1670. |
| 49 | Ventura M, Petrusek A, Miró A, Hamrová E, Buñay D, De Meester L, Mergeay J (2014) Local and regional founder effects in lake zooplankton persist after thousands of years despite high dispersal potential.Molecular Ecology, 23, 1014-1027. |
| 50 | Xu L (徐磊), Li SJ (李思嘉), Wang S (王晟), Han XY (韩小玉), Han BP (韩博平) (2014) Comparison of two methods for extracting DNA of resting eggs by Cladocera.Journal of Lake Sciences(湖泊科学), 26, 632-636. (in Chinese with English abstract) |
| 51 | Xu L (徐磊) (2013) Biogeography and Genetic Diversity of Two Cladoceran Species (Leptodora kindtii and Daphnia galeata) (两种枝角类(Leptodora kindtii 和 Daphnia galeata)的生物地理学及其种群遗传多样性研究). PhD dissertation, Jinan University, Guangzhou. (in Chinese with English abstract) |
| 52 | Zhang JC (张继承), Jiang QG (姜琦刚), Li YH (李远华), Wang K (王坤) (2008) Dynamic monitoring and climatic background of lake changes in Tibet based on RS/GIS.Journal of Earth Sciences and Environment(地球科学与环境学报), 30, 285-292. (in Chinese with English abstract) |
| [1] | Hong Deyuan. A brief discussion on methodology in taxonomy [J]. Biodiv Sci, 2025, 33(2): 24541-. |
| [2] | Wei Yi, Yi Ai, Meng Wu, Liming Tian, Tserang Donko Mipam. Soil archaeal community responses to different grazing intensities in the alpine meadows of the Qinghai-Tibetan Plateau [J]. Biodiv Sci, 2025, 33(1): 24339-. |
| [3] | Yajun Sun. What do higher or lower organisms mean—Clarify the meaning and validity of the biological ladder implied by On the Origin of Species [J]. Biodiv Sci, 2025, 33(1): 24394-. |
| [4] | Hua He, Dunyan Tan, Xiaochen Yang. Cryptic dioecy in angiosperms: Diversity, phylogeny and evolutionary significance [J]. Biodiv Sci, 2024, 32(6): 24149-. |
| [5] | Yanyu Ai, Haixia Hu, Ting Shen, Yuxuan Mo, Jinhua Qi, Liang Song. Vascular epiphyte diversity and the correlation analysis with host tree characteristics: A case in a mid-mountain moist evergreen broad-leaved forest, Ailao Mountains [J]. Biodiv Sci, 2024, 32(5): 24072-. |
| [6] | Yanwen Lv, Ziyun Wang, Yu Xiao, Zihan He, Chao Wu, Xinsheng Hu. Advances in lineage sorting theories and their detection methods [J]. Biodiv Sci, 2024, 32(4): 23400-. |
| [7] | Chen-Kun Jiang, Wen-Bin Yu, Guang-Yuan Rao, Huaicheng Li, Julien B. Bachelier, Hartmut H. Hilger, Theodor C. H. Cole. Plant Phylogeny Posters—An educational project on plant diversity from an evolutionary perspective [J]. Biodiv Sci, 2024, 32(11): 24210-. |
| [8] | Jiajia Chen, Zhen Pu, Zhonghong Huang, Fengqin Yu, Jianjun Zhang, Donghua Xu, Junquan Xu, Peng Shang, Dilimulati·Parhati, Yaojiang Li, Jigme Tshering, Yumin Guo. Global distribution and number of overwintering black-necked crane (Grus nigricollis) [J]. Biodiv Sci, 2023, 31(6): 22400-. |
| [9] | Xiaocheng Chen, Pengzhan Zhang, Bin Kang, Linshan Liu, Liang Zhao. Species and functional diversity of the passerine birds in the Tibetan Plateau based on specimens from the collection of Northwest Institute of Plateau Biology, Chinese Academy of Sciences [J]. Biodiv Sci, 2023, 31(5): 22638-. |
| [10] | Helu Zhang, Meihong Zhao, Shichun Sun, Xiaoshou Liu. Diversity and community characteristics of free-living nematodes in plateau salt lakes in Nagqu City, Tibet [J]. Biodiv Sci, 2023, 31(5): 22533-. |
| [11] | Xinyu Gong, Baorong Huang. Public welfare evaluation index system of national parks: A case study of the Qinghai-Tibet Plateau National Park Cluster [J]. Biodiv Sci, 2023, 31(3): 22571-. |
| [12] | Zhizhong Li, Shuai Peng, Qingfeng Wang, Wei Li, Shichu Liang, Jinming Chen. Cryptic diversity of the genus Ottelia in China [J]. Biodiv Sci, 2023, 31(2): 22394-. |
| [13] | Huiyin Song, Zhengyu Hu, Guoxiang Liu. Assessing advances in taxonomic research on Chlorellaceae (Chlorophyta) [J]. Biodiv Sci, 2023, 31(2): 22083-. |
| [14] | Dong Wang, Qinggaowa Sai, Zihan Wang, Hongxiu Zhao, Xinming Lian. Spatiotemporal overlap among sympatric Pallas’s cat (Otocolobus manul), Tibetan fox (Vulpes ferrilata) and red fox (V. vulpes) in the source region of the Yangtze River [J]. Biodiv Sci, 2022, 30(9): 21365-. |
| [15] | Ting Wang, Jiangping Shu, Yufeng Gu, Yanqing Li, Tuo Yang, Zhoufeng Xu, Jianying Xiang, Xianchun Zhang, Yuehong Yan. Insight into the studies on diversity of lycophytes and ferns in China [J]. Biodiv Sci, 2022, 30(7): 22381-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()