



Biodiv Sci ›› 2010, Vol. 18 ›› Issue (3): 262-274. DOI: 10.3724/SP.J.1003.2010.262 cstr: 32101.14.SP.J.1003.2010.262
• Special Issue • Previous Articles Next Articles
Xiying Ku1,2, Chuanjiang Zhou1,3, Shunping He1,*( )
)
Received:2009-10-29
Accepted:2010-01-01
Online:2010-05-20
Published:2012-02-08
Contact:
Shunping He
Xiying Ku, Chuanjiang Zhou, Shunping He. Validity of Pseudobagrus sinensis and mitochondrial DNA diversity of Pseudobagrus fulvidraco populations in China[J]. Biodiv Sci, 2010, 18(3): 262-274.
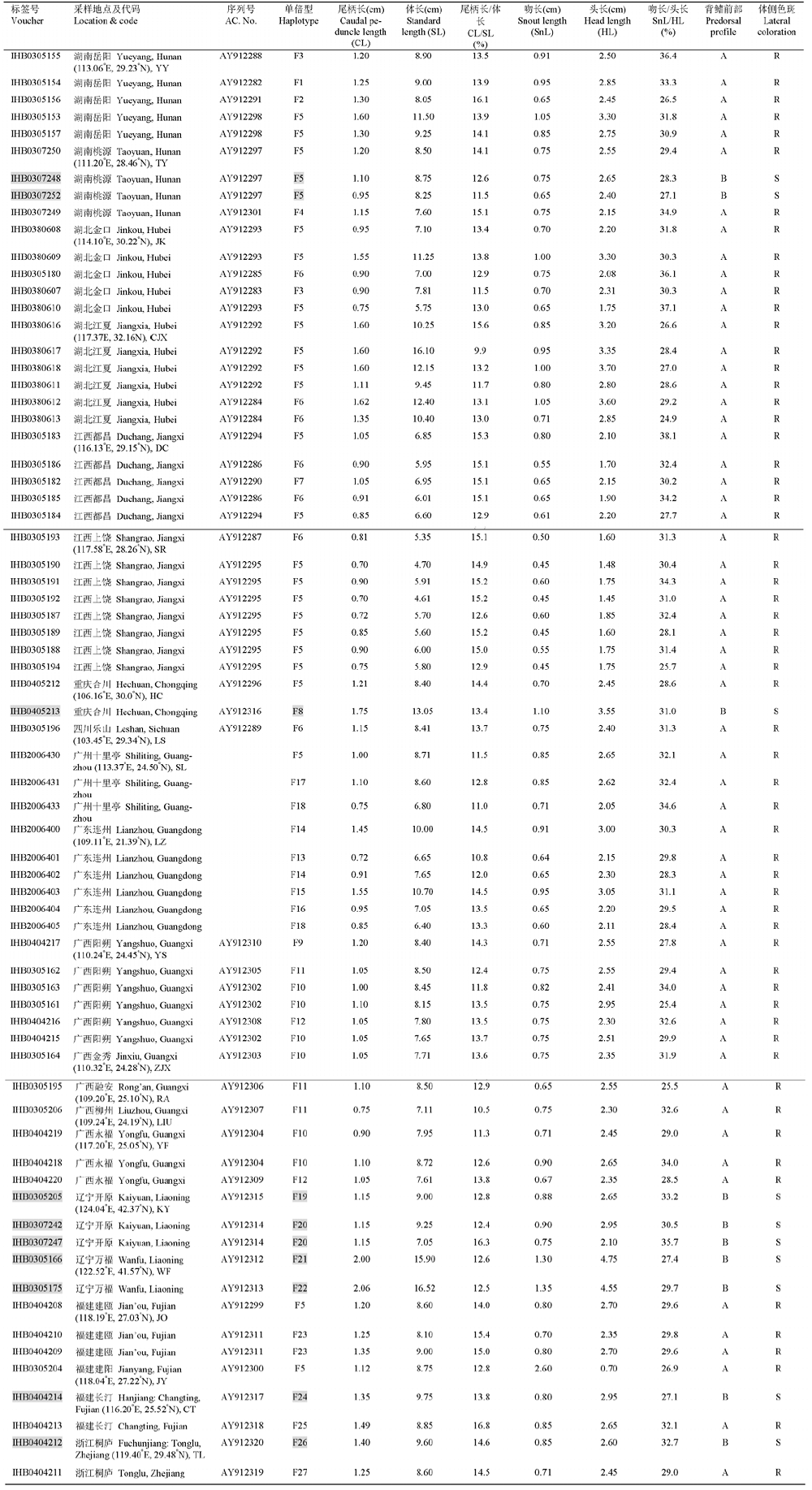 |
Table 1 Sampling and morphometric data for fish materials of Pseudobagrus fulvidraco and P. sinensis.Specimens under grey represent those provisionally identified as P. sinensis according to Ng & Kottelat ( 2007).
 |
| 尾柄长/体长比例 Caudal peduncle length / standard length | 吻长与/头长比例 Snout length / head length | ||||
|---|---|---|---|---|---|
| P. sinensis | P. fulvidraco | P. sinensis | P. fulvidraco | ||
| Ng & Kottelat ( | 9.5-10.9% | 7.3-9.6% | 29.8-32.2% | 32.2-37.5% | |
| 本研究测量结果 Measurements in this study | 11.5-16.3% | 9.9-16.8% | 27.1-35.7% | 24.9-38.1% | |
Table 2 Comparison of caudal peduncle and snout length between Pseudobagrus sinensisand P. fulvidraco
| 尾柄长/体长比例 Caudal peduncle length / standard length | 吻长与/头长比例 Snout length / head length | ||||
|---|---|---|---|---|---|
| P. sinensis | P. fulvidraco | P. sinensis | P. fulvidraco | ||
| Ng & Kottelat ( | 9.5-10.9% | 7.3-9.6% | 29.8-32.2% | 32.2-37.5% | |
| 本研究测量结果 Measurements in this study | 11.5-16.3% | 9.9-16.8% | 27.1-35.7% | 24.9-38.1% | |
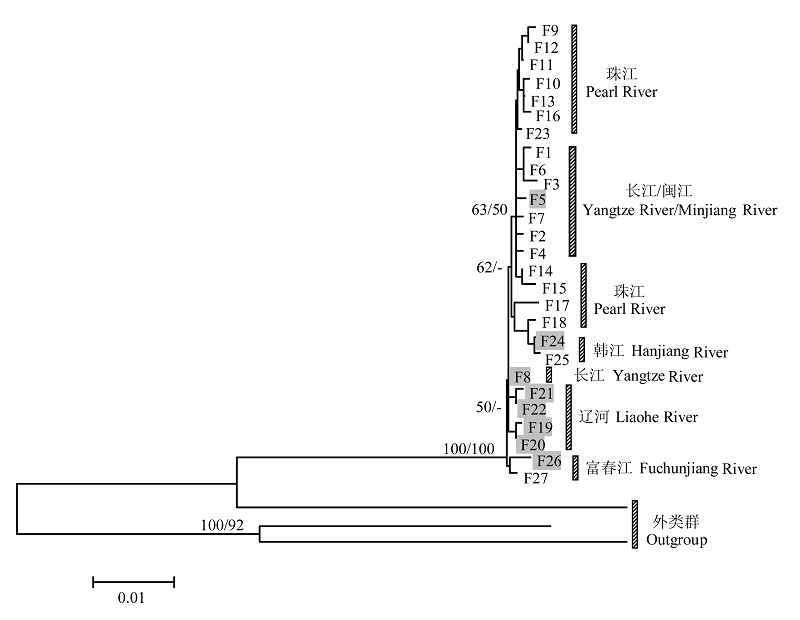
Fig. 1 The neighbor-joining (NJ) / maximum parsimony (MP) tree based on cyt bgene sequences. Values above nodes are proportions of 1,000 bootstrap pseudoreplicates in which the node was recovered in NJ/MP analysis (only values above 50% are shown). The grey boxes are haplotypes of Pseudobagrus sinensis.
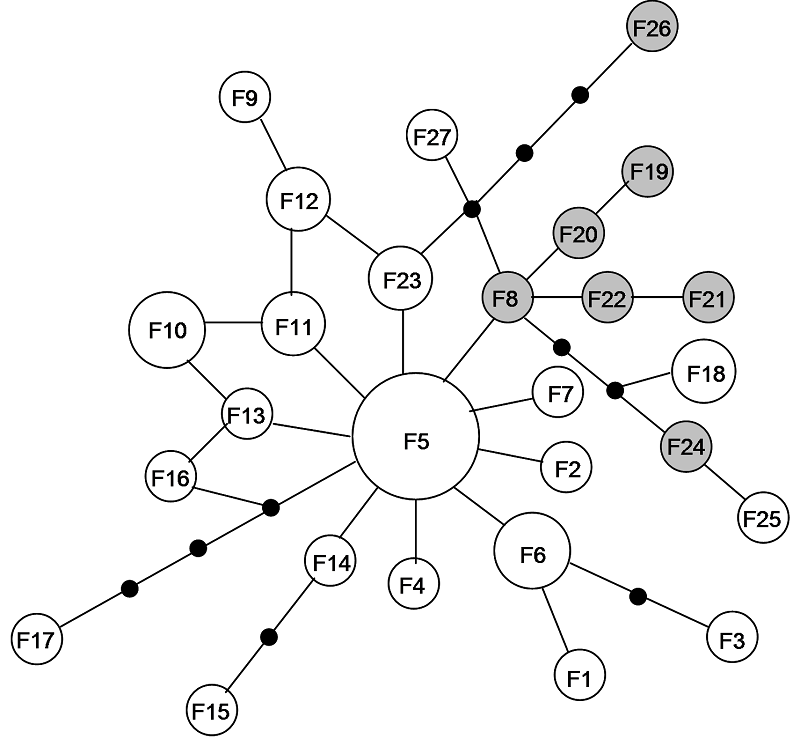
Fig. 2 Network relationships of haplotypes. Size of circle is proportional to the sample size of haplotypes. Small black circles represent a haplotype which is a necessary intermediate but is not found in the sampled population. Each line connecting haplotypes represent one mutation step. The grey circles are haplotypes of Pseudobagrus sinensis (F5 is not marked).
| 遗传变异来源 Source of variation | ΦST | ΦSC | ΦCT | 组间 Among groups (%) | 群体间 Within populations (%) |
|---|---|---|---|---|---|
| 所有群体 All populations | 0.892 | - | - | 89.15 | 10.85 |
| 水系间 Among drainages | 0.922 | 0.159 | 0.907 | 90.74 | 7.79 |
| 系统树的分支 Among clades | 0.959 | 0.607 | 0.897 | 89.66 | 4.07 |
| 物种间 Between species (P. sinensis/P. fulvidraco) | -0.147 | -0.562 | 0.265 | 26.5 | -41.26 |
Table 3 Hierarchical analysis of AMOVA
| 遗传变异来源 Source of variation | ΦST | ΦSC | ΦCT | 组间 Among groups (%) | 群体间 Within populations (%) |
|---|---|---|---|---|---|
| 所有群体 All populations | 0.892 | - | - | 89.15 | 10.85 |
| 水系间 Among drainages | 0.922 | 0.159 | 0.907 | 90.74 | 7.79 |
| 系统树的分支 Among clades | 0.959 | 0.607 | 0.897 | 89.66 | 4.07 |
| 物种间 Between species (P. sinensis/P. fulvidraco) | -0.147 | -0.562 | 0.265 | 26.5 | -41.26 |
| 长江 Yangtze River | 珠江 Pearl River | 闽江 Minjiang | 韩江 Hanjiang | 富春江 Fuchunjiang | 水系内不同群体间 Among groups within drainages | |
|---|---|---|---|---|---|---|
| 长江 Yangtze River | 0.0008 | |||||
| 珠江 Pearl River | 0.0040 | 0.0027 | ||||
| 闽江 Minjiang River | 0.0028 | 0.0050 | 0.0018 | |||
| 韩江 Hanjiang River | 0.0051 | 0.0067 | 0.0058 | 0.0006 | ||
| 富春江 Fuchunjiang River | 0.0035 | 0.0055 | 0.0038 | 0.0042 | 0.0009 | |
| 辽河 Liaohe River | 0.0058 | 0.0070 | 0.0068 | 0.0071 | 0.0059 | 0.0037 |
Table 4 Uncorrected p distances among groups of different drainages
| 长江 Yangtze River | 珠江 Pearl River | 闽江 Minjiang | 韩江 Hanjiang | 富春江 Fuchunjiang | 水系内不同群体间 Among groups within drainages | |
|---|---|---|---|---|---|---|
| 长江 Yangtze River | 0.0008 | |||||
| 珠江 Pearl River | 0.0040 | 0.0027 | ||||
| 闽江 Minjiang River | 0.0028 | 0.0050 | 0.0018 | |||
| 韩江 Hanjiang River | 0.0051 | 0.0067 | 0.0058 | 0.0006 | ||
| 富春江 Fuchunjiang River | 0.0035 | 0.0055 | 0.0038 | 0.0042 | 0.0009 | |
| 辽河 Liaohe River | 0.0058 | 0.0070 | 0.0068 | 0.0071 | 0.0059 | 0.0037 |
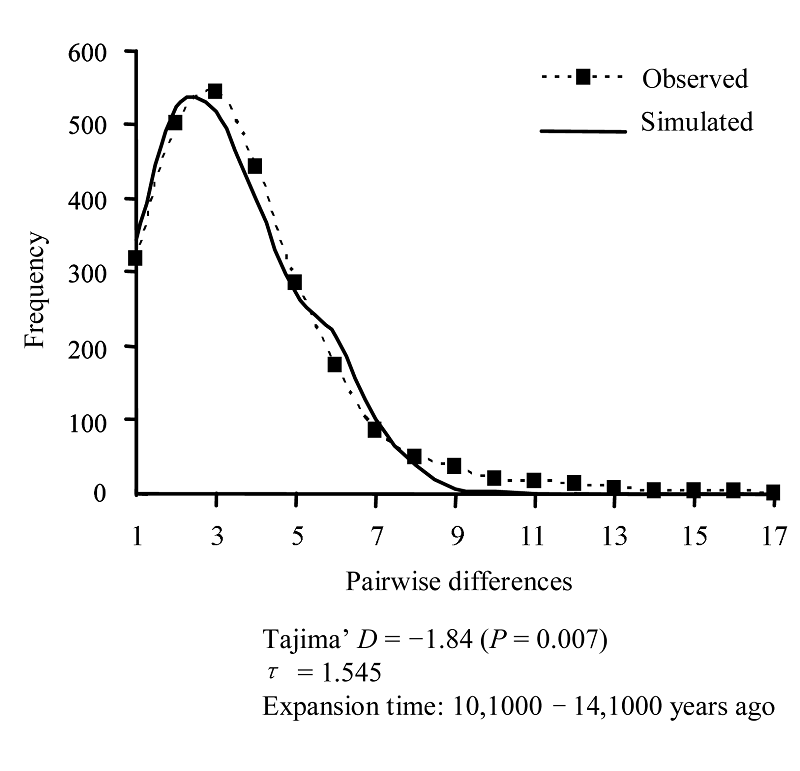
Fig. 3 Mismatch distribution for the population of Pseudobagrus fulvidraco. The solid line depicts the simulated mismatch distribution, the dashed line describes the observed distribution.
| [1] | Avise JC (2000) Empirical intraspecific phylogeography. In: Phylogeography, the History and Formation of Species. Harvard University Press, Cambridge, Massachusetts. |
| [2] | Avise JC, Walker D, Johns GC (1998) Speciation durations and Pleistocene effects on vertebrate phylogeography. Proceedings of the Royal Society of London, Series B, Biological Sciences, 265, 1707-1712. |
| [3] | Chu XL (褚新洛), Zheng BS (郑葆珊), Dai DY (戴定远) (1999) Fauna Sinica (Teleostei): Siluriformes (中国动物志: 硬骨鱼纲·鲇形目). Science Press, Beijing. (in Chinese) |
| [4] | Clark MK, Schoenbohm LM, Royden LH, Whipple KX, Burchfiel BC, Zhang X, Tang W, Wang E, Chen L (2004) Surface uplift, tectonics, and erosion of eastern Tibet from large-scale drainage patterns. Tectonics, 23, TC1006, doi: 10.1029/2002TC001402. |
| [5] |
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology, 9, 1657-1659.
DOI URL PMID |
| [6] | Compilatory Commission of Physical Geography of China, Chinese Academy of Sciences (中国科学院自然地理编委会) (1980) Physical Geography of China: Physiognomy (中国自然地理: 地貌). Science Press, Beijing. (in Chinese) |
| [7] |
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics (Online), 1, 47-50.
URL PMID |
| [8] | Fang YL (方耀林), Wang DQ (汪登强), Liu SP (刘绍平), Wu G (伍刚), Liao FC (廖伏初), Chen DQ (陈大庆) (2005) Variation in mitochondrial DNA of Pelteobagrus fulvidraco from three lakes in the Middle Yangtze River. Journal of Fishery Sciences of China (中国水产科学), 12, 56-61. (in Chinese with English abstract) |
| [9] | Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426. |
| [10] |
Hewitt GM (2000) The genetic legacy of the quaternary ice ages. Nature, 405, 907-913.
DOI URL PMID |
| [11] |
Hudson RR, Slatkint M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics, 132, 583-589.
URL PMID |
| [12] | Ku XY, Peng ZG, Diogo R, He SP (2007) MtDNA phylogeny provides evidence of generic polyphyleticism for East Asian bagrid catfishes. Hydrobiologia, 579, 147-159. |
| [13] | Lee CL, Kim IS (1990) A taxonomic revision of the family Bagridae (Pisces, Siluriformes) from Korea. Korean Journal of Ichthyology, 2, 117-137. |
| [14] | Liu HZ (刘焕章) (1998) A preliminary analysis to biogeographical process of the eastern Asian freshwater fishes. Acta Zootaxonomica Sinica (动物分类学报), 23, 49-55. (in Chinese with English abstract) |
| [15] | Liu SP (刘世平) (1997) A study on the biology of Pelteobagrus fulvidraco in Poyang Lake. Chinese Journal of Zoology (动物学杂志), 32 (4),10-16. (in Chinese with English abstract) |
| [16] | Lydeard C, Roe KJ (1997) The phylogenetic utility of the mitochondrial cytochrome b gene for inferring relationships among Actinopterygian fishes. In: Molecular Systematics of Fishes (eds Kocher TD, Stepien CA),pp.285-303. Academic Press, San Diego. |
| [17] |
Muse SV, Weir BS (1992) Testing for equality of evolutionary rates. Genetics, 132, 269-276.
URL PMID |
| [18] | Naseka AM, Bogutskaya NG (2004) Contribution to taxonomy and nomenclature of freshwater fishes of the Amur drainage area and the Far East (Pisces, Osteichthyes). Zoosystematica Rossica, 12, 279-290. |
| [19] | Ng HH, Dodson JJ (1999) Morphological and genetic descriptions of a new species of catfish, Hemibagrus chrysops, from Sarawak, East Malaysia, with an assessment of phylogenetic relationships (Teleostei: Bagridae). The Raffles Bulletin of Zoology, 47, 45-57. |
| [20] | Ng HH, Kottelat M (2007) The identity of Tachysurus sinensis (La Cepede, 1803), with the designation of a neotype (Teleostei: Bagridae) and notes on the identity of T. fulvidraco (Richardson, 1845). Electronic Journal of Ichthyology, 2, 35-45. |
| [21] | Peng ZG, He SP, Zhang YG (2002) Mitochondrial cytochrome b sequence variations and phylogeny of East Asian bagrid catfishes. Progress in Natural Science, 12, 421-425. |
| [22] |
Perdices A, Cunha C, Coelho MM (2004) Phylogenetic structure of Zacco platypus (Teleostei, Cyprinidae) populations on the upper and middle Chang Jiang (=Yangtze) drainage inferred from cytochrome b sequences. Molecular Phylogenetics and Evolution, 31, 192-203.
DOI URL PMID |
| [23] |
Perdices A, Sayanda D, Coelho MM (2005) Mitochondrial diversity of Opsariichthys bidens (Teleostei, Cyprinidae) in three Chinese drainages. Molecular Phylogenetics and Evolution, 37, 920-927.
DOI URL PMID |
| [24] |
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution, 49, 1280-1283.
DOI URL PMID |
| [25] | Ren ME (任美锷), Bao HS (包浩生), Han TC (韩同春) (1959) The Jinsha River valley landforms and river-capture in northwestern Yunnan. Acta Geographica Sinica (地理学报), 25 (2),135-155. (in Chinese) |
| [26] |
Rogers AR, Harpending A (1992) Population growth makes waves in distribution of pairwise genetic differences. Molecular Biology and Evolution, 9, 552-569.
DOI URL PMID |
| [27] | Song P (宋平), Pan YF (潘云风), Xiang Z (向筑), Hu JR (胡珈瑞), Hu YC (胡隐昌), Cai CL (蔡从利) (2001) RAPD markers and genetic diversity in Pelteobagrus fulvidraco. Journal of Wuhan University (Natural Science Edition) (武汉大学学报自然科学版), 47, 233-237. (in Chinese with English abstract) |
| [28] | Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts. |
| [29] |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595.
URL PMID |
| [30] |
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599.
DOI URL PMID |
| [31] |
Thompson JD, Gibson TJ, Plewnink F (1997) The Clustal_X windows interface: flexible strategies for multiple sequences alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882.
DOI URL PMID |
| [32] |
Wang JP, Hsu KC, Chiang TY (2000) Mitochondrial DNA phylogeography of Acrossocheilus paradoxus (Cyprinidae) in Taiwan. Molecular Ecology, 9, 1483-1494.
DOI URL PMID |
| [33] |
Wang ZW, Wu QJ, Zhou JF, Ye YZ (2004) Geographic distribution of Pelteobagrus fulvidraco and Pelteobagrus vachelli in the Yangtze River based on mitochondrial DNA markers. Biochemical Genetics, 42, 391-400.
DOI URL PMID |
| [34] | Watanabe K, Nishida M (2003) Genetic population structure of Japanese bagrid catfishes. Ichthyological Research, 50, 140-148. |
| [35] |
Yang L, He SP (2008) Phylogeography of the freshwater catfish Hemibagrus guttatus (Siluriformes, Bagridae): implications for South China biogeography and influence of sea-level changes. Molecular Phylogenetics and Evolution, 49, 393-398.
DOI URL PMID |
| [36] |
Yang L, Mayden RL, He SP (2009) Population genetic structure and geographical differentiation of the Chinese catfish Hemibagrus macropterus (Siluriformes, Bagridae): evidence for altered drainages patterns. Molecular Phylogenetics and Evolution, 51, 405-411.
DOI URL PMID |
| [37] | Yap SY (2002) On the distributional patterns of Southeast-East Asian freshwater fish and their history. Journal of Biogeography, 29, 1187-1199. |
| [38] | Zhang Y (张燕), Zhang E (张鹗), He SP (何舜平) (2003) Studies on the structure of the control region of the Bagridae in China and its phylogenetic significance. Acta Hydrobiologica Sinica (水生生物学报), 27, 463-467. (in Chinese with English abstract) |
| [39] | Zhao K, Duan ZY, Yang GS, Peng ZG, He SP, Chen YY (2007) Origin of Gymnocypris przewalskii and phylogenetic history of Gymnocypris eckloni (Teleostei: Cyprinidae). Progress in Natural Science, 17, 520-528. |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Yanwen Lv, Ziyun Wang, Yu Xiao, Zihan He, Chao Wu, Xinsheng Hu. Advances in lineage sorting theories and their detection methods [J]. Biodiv Sci, 2024, 32(4): 23400-. |
| [3] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [4] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [5] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [6] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [7] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [8] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [11] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [12] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [13] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [14] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [15] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()