



Biodiv Sci ›› 2013, Vol. 21 ›› Issue (5): 601-609. DOI: 10.3724/SP.J.1003.2013.09098 cstr: 32101.14.SP.J.1003.2013.09098
• Orginal Article • Previous Articles Next Articles
Deyun Wang1, Jie Peng1, Yajing Chen1, Guosheng Lü1, Xiaoping Zhang1,2, Jianwen Shao1,3,*( )
)
Received:2013-04-18
Accepted:2013-07-01
Online:2013-09-20
Published:2013-10-08
Contact:
Shao Jianwen
Deyun Wang,Jie Peng,Yajing Chen,Guosheng Lü,Xiaoping Zhang,Jianwen Shao. Genetic diversity and genetic structure of the rare and endangered species, Primula ranunculoides[J]. Biodiv Sci, 2013, 21(5): 601-609.
| 种群 Population | 地点 Locality | 生境 Habitat | 地理位置 Geographical location | 海拔 Altitude (m) | 面积 Area (m2) | 花朵类型 Flower type | 种群大小 Population size |
|---|---|---|---|---|---|---|---|
| 杨家坪A YJPA | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside under forests | 28°47'45"N, 114°43'56"E | 300-375 | 720 | 两型花 Distyle | ~2,000 |
| 杨家坪B YJPB | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°47'04"N, 114°44'13"E | 350-360 | 1,060 | 两型花 Distyle | ~6,000 |
| 毛竹山 MZS | 江西省修水县毛竹山 Maozhushan, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°50'31"N, 114°52'00"E | 390-410 | 1,650 | 两型花 Distyle | ~1,500 |
| 三界A SJA | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°24'16"N, 114°27'15"E | 255-285 | 545 | 两型花 Distyle | ~1,000 |
| 三界B SJB | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°22'57"N, 114°27'08"E | 265-280 | 385 | 两型花 Distyle | ~700 |
| 七姊妹山 JPT | 湖北省宣恩县七姊妹山 Qizimei Mount, Xuanen County, Hubei | 林下溪边 Brook-side, under forest | 30°02'09"N, 109°43'57"E | 1,310-1,365 | 1,200 | 同型花 Monostyle | ~600 |
| 银炉 YL | 江西省武宁县银炉 Yinlu, Wuning County, Jiangxi | 溪边石壁 Rock near brook-side | 28°59'31''N, 114°49'39''E | 420 | 50 | 两型花 Distyle | ~200 |
Table 1 Locality and information of studied populations of Primula ranunculoides
| 种群 Population | 地点 Locality | 生境 Habitat | 地理位置 Geographical location | 海拔 Altitude (m) | 面积 Area (m2) | 花朵类型 Flower type | 种群大小 Population size |
|---|---|---|---|---|---|---|---|
| 杨家坪A YJPA | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside under forests | 28°47'45"N, 114°43'56"E | 300-375 | 720 | 两型花 Distyle | ~2,000 |
| 杨家坪B YJPB | 江西省修水县杨家坪 Yangjiaping, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°47'04"N, 114°44'13"E | 350-360 | 1,060 | 两型花 Distyle | ~6,000 |
| 毛竹山 MZS | 江西省修水县毛竹山 Maozhushan, Xiushui County, Jiangxi | 林下溪边或路边 Brook-side or roadside, under forest | 28°50'31"N, 114°52'00"E | 390-410 | 1,650 | 两型花 Distyle | ~1,500 |
| 三界A SJA | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°24'16"N, 114°27'15"E | 255-285 | 545 | 两型花 Distyle | ~1,000 |
| 三界B SJB | 湖北省通山县三界 Sanjie, Tongshan County, Hubei | 林下溪边或路边 Brook-side or roadside, under forest | 29°22'57"N, 114°27'08"E | 265-280 | 385 | 两型花 Distyle | ~700 |
| 七姊妹山 JPT | 湖北省宣恩县七姊妹山 Qizimei Mount, Xuanen County, Hubei | 林下溪边 Brook-side, under forest | 30°02'09"N, 109°43'57"E | 1,310-1,365 | 1,200 | 同型花 Monostyle | ~600 |
| 银炉 YL | 江西省武宁县银炉 Yinlu, Wuning County, Jiangxi | 溪边石壁 Rock near brook-side | 28°59'31''N, 114°49'39''E | 420 | 50 | 两型花 Distyle | ~200 |
| 位点 Locus | 序列 Repeat motif | 引物 Primer (5'-3') | 退火温度(℃) Annealing temperature | 荧光标记 Fluorescent label | 等位基因数 No. of alleles |
|---|---|---|---|---|---|
| Pm1 | (TC)10(CT)3 | F:ATCTTTGAGGTCCTTTTA | 50 | FAM | 7 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm2 | (AG)14 | F: CGCCTACAGTGTTTGGGA | 55 | FAM | 2 |
| R: CTATCTCACCTGCGTTCT | |||||
| Pm7 | (AG)3GG(AG)8 | F: TTGTTCACCGACGCATAC | 54 | HEX | 18 |
| R: TTACACGCACCAAATCAT | |||||
| Pm9 | (TTTC)2(TC)6 | F: AGACTCACGAGGAATACG | 50 | HEX | 6 |
| R: AGAAAAGGAGGAGACAAA | |||||
| Pm12 | (TC)10(CT)3 | F: TAAAACTCCTGGAGGGGTAC | 51 | TAMRA | 10 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm13 | (GATAGG)2GAT(AG)7 | F: GAGGACAGGCACCACAGA | 54 | TAMRA | 6 |
| R: TCCCCAACTTCATGCTCTT | |||||
| Pm14 | (AG)3(GA)12 | F: TCGCCCAATGGAGTGAAC | 50 | TAMRA | 9 |
| R: TCTTTGAGGTCCTTTTAT | |||||
| Pm16 | (GAGGGA)3(GA)3 | F: AACCACTCGTCGTCCTAA | 51 | FAM | 5 |
| R: CGATAGATTGCCTTACCC | |||||
| Pm17 | (GA)9 | F: TAAATCAAGGTAGCAACT | 51 | HEX | 9 |
| R: TACCTACCATTACTCCC |
Table 2 The sequences of microsatellite loci and primers in this study
| 位点 Locus | 序列 Repeat motif | 引物 Primer (5'-3') | 退火温度(℃) Annealing temperature | 荧光标记 Fluorescent label | 等位基因数 No. of alleles |
|---|---|---|---|---|---|
| Pm1 | (TC)10(CT)3 | F:ATCTTTGAGGTCCTTTTA | 50 | FAM | 7 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm2 | (AG)14 | F: CGCCTACAGTGTTTGGGA | 55 | FAM | 2 |
| R: CTATCTCACCTGCGTTCT | |||||
| Pm7 | (AG)3GG(AG)8 | F: TTGTTCACCGACGCATAC | 54 | HEX | 18 |
| R: TTACACGCACCAAATCAT | |||||
| Pm9 | (TTTC)2(TC)6 | F: AGACTCACGAGGAATACG | 50 | HEX | 6 |
| R: AGAAAAGGAGGAGACAAA | |||||
| Pm12 | (TC)10(CT)3 | F: TAAAACTCCTGGAGGGGTAC | 51 | TAMRA | 10 |
| R: ATCGCCCAATGGAGTGAA | |||||
| Pm13 | (GATAGG)2GAT(AG)7 | F: GAGGACAGGCACCACAGA | 54 | TAMRA | 6 |
| R: TCCCCAACTTCATGCTCTT | |||||
| Pm14 | (AG)3(GA)12 | F: TCGCCCAATGGAGTGAAC | 50 | TAMRA | 9 |
| R: TCTTTGAGGTCCTTTTAT | |||||
| Pm16 | (GAGGGA)3(GA)3 | F: AACCACTCGTCGTCCTAA | 51 | FAM | 5 |
| R: CGATAGATTGCCTTACCC | |||||
| Pm17 | (GA)9 | F: TAAATCAAGGTAGCAACT | 51 | HEX | 9 |
| R: TACCTACCATTACTCCC |
| 种群 Population | 取样数 Sample size | 有效种群大小 Effective population size | 期望杂合度(He) Expected heterozygosity | 观察杂合度(Ho) Observed heterozygosity | 稀有等位基因 Rare alleles | 近交系数(Fis) Inbreeding coefficient |
|---|---|---|---|---|---|---|
| YJPA | 32 | 166 | 0.430 | 0.424 | 2 | 0.014 ns |
| YJPB | 31 | 231 | 0.412 | 0.399 | 4 | 0.032** |
| MZS | 30 | 105 | 0.297 | 0.155 | 2 | 0.484*** |
| SJA | 33 | 169 | 0.348 | 0.330 | 4 | 0.055*** |
| SJB | 31 | 149 | 0.281 | 0.230 | 1 | 0.184*** |
| JPT | 33 | 127 | 0.203 | 0.132 | 2 | 0.350*** |
| YL | 32 | 153 | 0.340 | 0.328 | 2 | 0.037** |
| 平均 Mean | 31.7 | 157.1 | 0.330 | 0.286 | 2.4 |
Table 3 Genetic characteristics of Primula ranunculoides populations
| 种群 Population | 取样数 Sample size | 有效种群大小 Effective population size | 期望杂合度(He) Expected heterozygosity | 观察杂合度(Ho) Observed heterozygosity | 稀有等位基因 Rare alleles | 近交系数(Fis) Inbreeding coefficient |
|---|---|---|---|---|---|---|
| YJPA | 32 | 166 | 0.430 | 0.424 | 2 | 0.014 ns |
| YJPB | 31 | 231 | 0.412 | 0.399 | 4 | 0.032** |
| MZS | 30 | 105 | 0.297 | 0.155 | 2 | 0.484*** |
| SJA | 33 | 169 | 0.348 | 0.330 | 4 | 0.055*** |
| SJB | 31 | 149 | 0.281 | 0.230 | 1 | 0.184*** |
| JPT | 33 | 127 | 0.203 | 0.132 | 2 | 0.350*** |
| YL | 32 | 153 | 0.340 | 0.328 | 2 | 0.037** |
| 平均 Mean | 31.7 | 157.1 | 0.330 | 0.286 | 2.4 |
| YJPA | YJPB | MZS | SJA | SJB | JPT | YL | |
|---|---|---|---|---|---|---|---|
| YJPA | - | 0.091 | 0.208 | 0.420 | 0.528 | 0.875 | 0.702 |
| YJPB | 0.096 | - | 0.167 | 0.311 | 0.363 | 0.793 | 0.588 |
| MZS | 0.256 | 0.222 | - | 0.336 | 0.378 | 0.827 | 0.742 |
| SJA | 0.301 | 0.262 | 0.399 | - | 0.030 | 0.921 | 0.473 |
| SJB | 0.385 | 0.332 | 0.476 | 0.047 | - | 0.929 | 0.467 |
| JPT | 0.510 | 0.526 | 0.645 | 0.569 | 0.599 | - | 0.318 |
| YL | 0.445 | 0.424 | 0.536 | 0.392 | 0.428 | 0.367 | - |
Table 4 Pairwise population differentiation (below diagonal) and genetic distances (above diagonal) among Primula ranunculoides populations
| YJPA | YJPB | MZS | SJA | SJB | JPT | YL | |
|---|---|---|---|---|---|---|---|
| YJPA | - | 0.091 | 0.208 | 0.420 | 0.528 | 0.875 | 0.702 |
| YJPB | 0.096 | - | 0.167 | 0.311 | 0.363 | 0.793 | 0.588 |
| MZS | 0.256 | 0.222 | - | 0.336 | 0.378 | 0.827 | 0.742 |
| SJA | 0.301 | 0.262 | 0.399 | - | 0.030 | 0.921 | 0.473 |
| SJB | 0.385 | 0.332 | 0.476 | 0.047 | - | 0.929 | 0.467 |
| JPT | 0.510 | 0.526 | 0.645 | 0.569 | 0.599 | - | 0.318 |
| YL | 0.445 | 0.424 | 0.536 | 0.392 | 0.428 | 0.367 | - |
| 变异来源 Source | 自由度 d.f. | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例% Percentage of variation | 显著性检验 Significance tests |
|---|---|---|---|---|---|
| 种群间 Among populations | 6 | 225.635 | 0.6596 | 48.08 | P<0.001 |
| 种群内 Within populations | 215 | 310.851 | 0.7124 | 51.92 | P<0.001 |
| 总和 Total | 222 | 536.486 | 1.3720 | 100% |
Table 5 Analysis of molecular variance (AMOVA) within/among Primula ranunculoides populations
| 变异来源 Source | 自由度 d.f. | 方差和 Sum of squares | 变异组分 Variance components | 占总变异比例% Percentage of variation | 显著性检验 Significance tests |
|---|---|---|---|---|---|
| 种群间 Among populations | 6 | 225.635 | 0.6596 | 48.08 | P<0.001 |
| 种群内 Within populations | 215 | 310.851 | 0.7124 | 51.92 | P<0.001 |
| 总和 Total | 222 | 536.486 | 1.3720 | 100% |
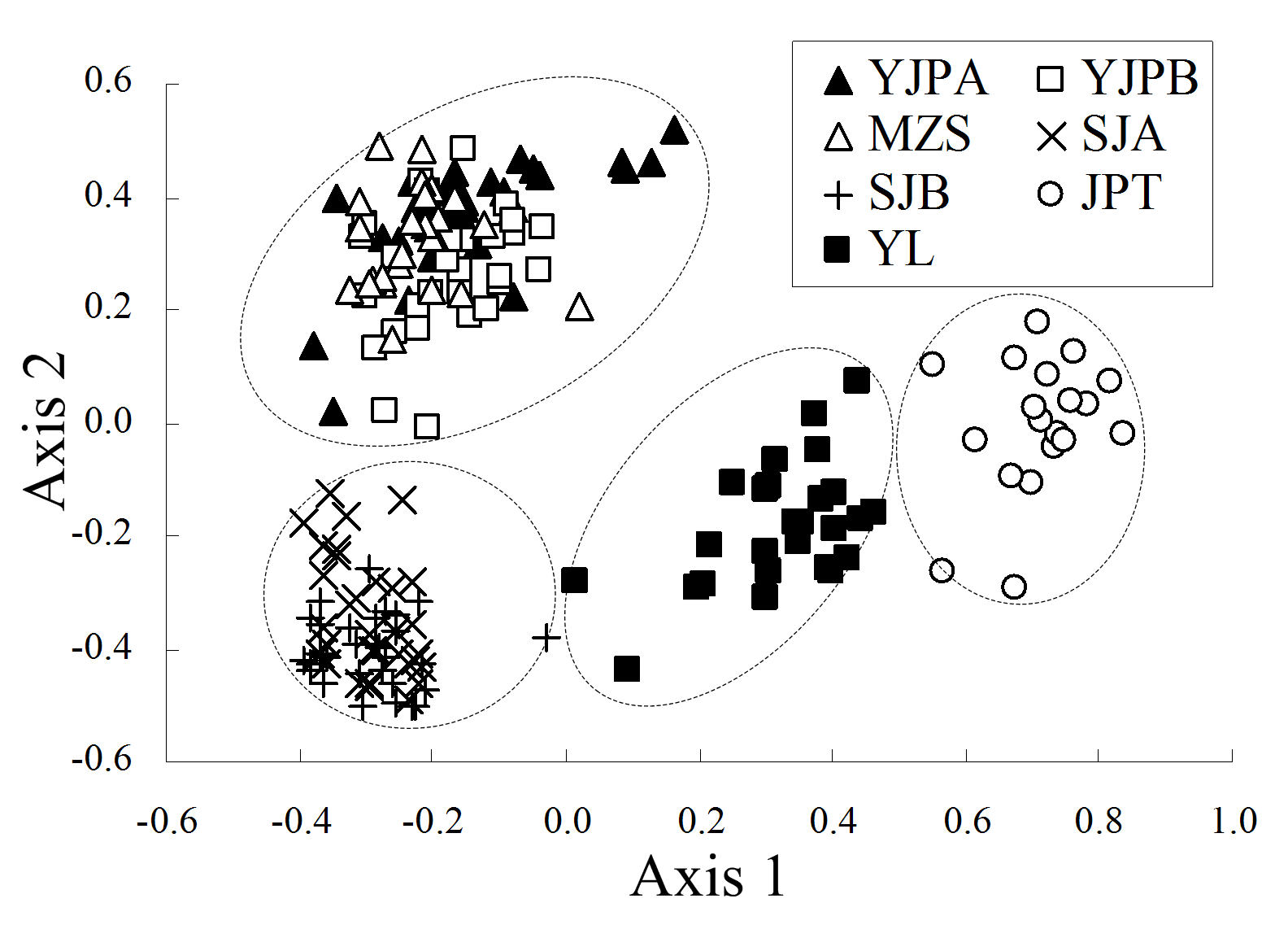
Fig. 2 The result of Principal coordinates (PCO) analysis of Primula ranunculoides populations. The first and second axis extracted 40.0% and 26.9% of the total genetic variance, respectively. Population codes see Table 1.
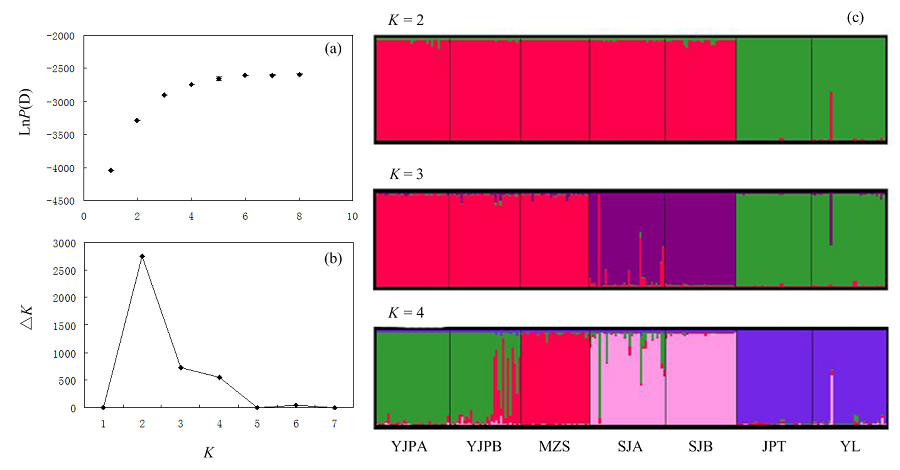
Fig. 3 The results of genetic structure analyses by STRUCTURE soft. (a) Plot of mean posterior probability lnP(D) values of each K; (b) The corresponding △K statistics calculated according to Evanno et al. (2005); (c) Histogram of the structure analysis for the model with K = 2-4. Population codes see Table 1.
| 1 | Beerli P (2008) MIGRATE Version 3.0―a Maximum Likelihood and Bayesian Estimator of Gene Flow Using the Coalescent. Distributed over the Internet at. |
| 2 | Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, Logiciel sous WindowsTM pour la Génétique des Populations. Laboratoire Génome, Populations, Intera- ctions, Université de Montpellier II, Montpellier. |
| 3 | Chen FH (1948) A new Chinese Primula.Notes from the Royal Botanic Garden Edinburgh, 20, 120. |
| 4 | Hu CM (胡启明) (1990) Primulaceae. In: Flora Reipublicae Popularis Sinicae (中国植物志) (ed. Wu ZY (吴征镒)), Tomus 59(2), pp. 1-277. Science Press, Beijing. (in Chinese) |
| 5 | Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation.Annual Review of Ecology and Systematics, 24, 217-242. |
| 6 | Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study.Molecular Ecology, 14, 2611-2620. |
| 7 | Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis.Evolutionary Bioinformatics Online, 1, 47-50. |
| 8 | Foxe JP, Stift M, Tedder A, Haudry A, Wright SI, Mable BK (2010) Reconstructing origins of loss of self-incompatibility and selfing in north American Arabidopsis lyrata: a population genetic context.Evolution, 64, 3495-3510. |
| 9 | Guillernsut P, Maréchal-Drouard L (1992) Isolation of plant DNA: a fast, inexpensive, and reliable method.Plant Molecular Biology Reporter, 10, 60-65. |
| 10 | Guillot G, Mortier F, Estoup A (2005) GENELAND: a computer package for landscape genetics.Molecular Ecology Notes, 5, 712-715. |
| 11 | Hao G, Hu CM, Lee NS (2002) Circumscriptions and phylogenetic relationships of Primula sect. Auganthus and Ranunculoides: evidence from nrDNA ITS sequences.Acta Botanica Sinica, 44, 72-75. |
| 12 | Honjo M, Kitamoto N, Ueno S, Tsumura Y, Washitani I, Ohsawa R (2009) Management units of the endangered herb Primula sieboldii based on microsatellite variation among and within populations throughout Japan.Conservation Genetics, 10, 257-267. |
| 13 | Hu CM, Kelso S (1996) Primulaceae. In: Flora of China (eds Wu ZY, Raven PH), Vol. 15, pp. 99-185. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis. |
| 14 | Huang Y, Wang XQ, Yang CY, Long CL (2010) Development of 11 polymorphic microsatellite loci from Primula amthystina Franchet. (Primulaceae).Hortscience, 45, 148-149. |
| 15 | Ishihama F, Ueno S, Tsumura Y, Washitani I (2006) Effects of density and floral morph on pollen flow and seed reproduction of an endangered heterostylous herb, Primula sieboldii.Journal of Ecology, 94, 846-855. |
| 16 | Jacquemyn H, Olivier H, Peter G, Isabel RU (2004) Genetic structure of the forest herb Primula elatior in a changing landscape.Molecular Ecology, 13, 211-219. |
| 17 | Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure.Bioinformatics, 23, 1801-1806. |
| 18 | Kaeuffer R, Réale D, Coltman DW, Pontier D (2007) Detecting population structure using Structure software: effect of background linkage disequilibrium.Heredity, 99, 374-380. |
| 19 | Lienert J, Fischer M (2003) Habitat fragmentation affects the common wetland specialist Primula farinosa in north-east Switzerland.Journal of Ecology, 91, 587-599. |
| 20 | Manni F, Guérard E (2004) BARRIER Version 2.2. Manual of the User. Population Genetics Team, Museum of Mankind, Paris. |
| 21 | Mantel N (1967) The detection of disease clustering and a generalized regression approach.Cancer Research, 27, 209-220. |
| 22 | Miller MP (1997) Tools for Population Genetic Analysis (TFPGA), Version 1.3. Department of Biological Sciences. Northern Arizona University, Arizona. |
| 23 | Ness RW, Wright SI, Barrett SCH (2010) Mating-system variation, demographic history and patterns of nucleotide diversity in the tristylous plant Eichhornia paniculata.Genetics, 184, 381-392. |
| 24 | Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants.Perspectives in Plant Ecology Evolution and Systematics, 3, 93-114. |
| 25 | Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research.Molecular Ecology Notes, 6, 288-295. |
| 26 | Peng YQ, Shao JW, Wu HL, Zhang XP, Zhu GP (2009) Isolation and characterization of fifteen polymorphic microsatellite loci from Primula merrilliana (Primulaceae), an endemic from China.Conservation Genetics, 10, 1441-1443. |
| 27 | Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data.Genetics, 155, 945-959. |
| 28 | Richards AJ (2003) Primula, 2nd edn. Timber Press, Portland. |
| 29 | Rosenberg NA (2004) Distruct: a program for the graphical display of population structure.Molecular Ecology Notes, 4, 137-138. |
| 30 | Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux.Molecular Ecology Resources, 8, 103-106. |
| 31 | Shao JW, Chen WL, Peng YQ, Zhu GP, Zhang XP (2009) Genetic diversity within and among populations of the endan gered and endemic species Primula merrilliana in China. Biochemical Systematics and Ecology, 37, 699-706. |
| 32 | Shao JW, Wu YF, Kan XZ, Liang TJ, Zhang XP (2012) Reappraisal of Primula ranunculoides (Primulaceae), an endangered species endemic to China, based on morphological, molecular genetic and reproductive characters.Botanical Journal of the Linnean Society, 169, 338-349. |
| 33 | Shao JW (邵剑文), Zhang XP (张小平), Zhang ZX (张中信), Zhu GP (朱国萍) (2008a) Identification of effective pollinators of Primula merrilliana and effects of flower density and population size on pollination efficiency.Journal of Systematics and Evolution, 46, 537-544. (in Chinese with English abstract) |
| 34 | Shao JW, Zhang ZX, Zhu GP, Zhang XP (2008b) Effects of population size on reproductive success of the endangered and endemic species Primula merrilliana. Journal of Integrative Plant Biology, 50, 1151-1160. |
| 35 | Slatkin M (1985) Gene flow in natural populations.Annual Review of Ecology and Systematics, 16, 393-430. |
| 36 | van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data.Molecular Ecology Notes, 4, 535-538. |
| 37 | van Rossum F, De Sousa SC, Triest L (2004) Genetic consequences of habitat fragmentation in an agricultural landscape on the common Primula veris, and comparison with its rare congener, P. vulgaris.Conservation Genetics, 5, 231-245. |
| 38 | van Rossum F, Triest L (2003) Spatial genetic structure and reproductive success in fragmented and continuous populations of Primula vulgaris.Folia Geobotanica, 38, 239-254. |
| 39 | van Rossum F, Triest L (2006) Fine-scale genetic structure of the common Primula elatior (Primulaceae) at an early stage of population fragmentation.American Journal of Botany, 93, 1281-1288. |
| 40 | Xue DW, Ge XJ, Hao G, Zhang CQ (2004) High genetic diversity in a rare, narrowly endemic primrose species: Primula interjacens by ISSR analysis.Acta Botanica Sinica, 46, 1163-1169. |
| 41 | Yan HF, Ge XJ, Hu CM, Hao G (2010) Isolation and characterization of microsatellite loci for the ornamental plant Primula obconica Hance (Primulaceae).Hortscience, 45, 314-315. |
| 42 | Yuan N, Comes HP, Mao YR, Qi XS, Qiu YX (2012) Genetic effects of recent habitat fragmentation in the Thousand-Island Lake region of southeast China on the distylous herb Hedyotis chrysotricha (Rubiaceae).American Journal of Botany, 99, 1715-1725. |
| 43 | Zhang DX, Hewitt GM (2003) Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects.Molecular Ecology, 12, 563-584. |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [3] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [4] | Xianglin Yang, Caiyun Zhao, Junsheng Li, Fangfang Chong, Wenjin Li. Invasive plant species lead to a more clustered community phylogenetic structure: An analysis of herbaceous plants in Guangxi’s national nature reserves [J]. Biodiv Sci, 2024, 32(11): 24175-. |
| [5] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [6] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [7] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [8] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [9] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [10] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [11] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [12] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [13] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [14] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [15] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()