



Biodiv Sci ›› 2013, Vol. 21 ›› Issue (3): 306-314. DOI: 10.3724/SP.J.1003.2013.09029 cstr: 32101.14.SP.J.1003.2013.09029
• Orginal Article • Previous Articles Next Articles
Jingjing Li1,2, Jie Zhang1,2, Zimin Hu1,*( ), Delin Duan1,*(
), Delin Duan1,*( )
)
Received:2013-01-30
Accepted:2013-03-28
Online:2013-05-20
Published:2013-06-05
Contact:
Hu Zimin,Duan Delin
Jingjing Li,Jie Zhang,Zimin Hu,Delin Duan. Population genetics and demographic history of red seaweed, Palmaria palmata, from the Canada-northwest Atlantic[J]. Biodiv Sci, 2013, 21(3): 306-314.
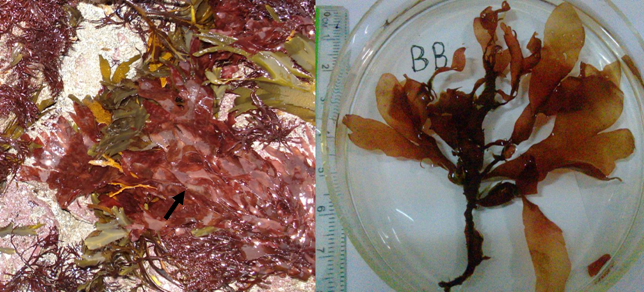
Fig. 1 Morphological characters of Palmaria palmata. (a) Living intertidal habitat of P. palmata from County Clare, Ireland in 2008. It is frequently found living on thallus of red marcroalga Chondrus crispus and brown macroalga Fucus serratus; (b) Purple morphological features of P. palmata from Bonne Bay, Canada.
| 简称 Code | 种群 Population | 经纬度 Locality | N | Nh | H | h (SD) | π (SD) |
|---|---|---|---|---|---|---|---|
| Gulf of St. Lawrence | |||||||
| LA | L’Anse Amour, Canada | 52.32°N, 56.60°W | 25 | 3 | C1, C2, C3 | 0.640 (0.052) | 0.0024 (0.0020) |
| BB | Bonne Bay, Canada | 49.51°N, 57.92°W | 31 | 3 | C1, C3, C4 | 0.540 (0.043) | 0.0018 (0.0017) |
| GP | Gaspe, Canada | 48.88°N, 64.50°W | 11 | 1 | C3 | 0.000 (0.000) | 0.0000 (0.0000) |
| RS | Rimouski, Canada | 48.47°N, 68.51°W | 24 | 2 | C3, C5 | 0.083 (0.075) | 0.0002 (0.0005) |
| Bay of Fundy | |||||||
| MB | Maces Bay, Canada | 45.11°N, 66.75°W | 8 | 2 | C3, C6 | 0.250 (0.180) | 0.0008 (0.0011) |
| VB | Victoria Beach, Canada | 44.69°N, 65.75°W | 9 | 3 | C3, C7, C8 | 0.417 (0.191) | 0.0014 (0.0016) |
| LT | Letete, Canada | 45.06°N, 66.89°W | 12 | 2 | C3, C9 | 0.303 (0.148) | 0.0009 (0.0012) |
| WH | White Head, Canada | 44.63°N, 66.72°W | 18 | 3 | C3, C10, C11 | 0.451 (0.117) | 0.0015 (0.0015) |
Table 1 Sampling details of eight Palmaria palmata populations, including abbreviation codes, geographic location, sample size (N), number of haplotypes (Nh), types of haplotype (H), haplotype diversity (h), and nucleotide diversity (π)
| 简称 Code | 种群 Population | 经纬度 Locality | N | Nh | H | h (SD) | π (SD) |
|---|---|---|---|---|---|---|---|
| Gulf of St. Lawrence | |||||||
| LA | L’Anse Amour, Canada | 52.32°N, 56.60°W | 25 | 3 | C1, C2, C3 | 0.640 (0.052) | 0.0024 (0.0020) |
| BB | Bonne Bay, Canada | 49.51°N, 57.92°W | 31 | 3 | C1, C3, C4 | 0.540 (0.043) | 0.0018 (0.0017) |
| GP | Gaspe, Canada | 48.88°N, 64.50°W | 11 | 1 | C3 | 0.000 (0.000) | 0.0000 (0.0000) |
| RS | Rimouski, Canada | 48.47°N, 68.51°W | 24 | 2 | C3, C5 | 0.083 (0.075) | 0.0002 (0.0005) |
| Bay of Fundy | |||||||
| MB | Maces Bay, Canada | 45.11°N, 66.75°W | 8 | 2 | C3, C6 | 0.250 (0.180) | 0.0008 (0.0011) |
| VB | Victoria Beach, Canada | 44.69°N, 65.75°W | 9 | 3 | C3, C7, C8 | 0.417 (0.191) | 0.0014 (0.0016) |
| LT | Letete, Canada | 45.06°N, 66.89°W | 12 | 2 | C3, C9 | 0.303 (0.148) | 0.0009 (0.0012) |
| WH | White Head, Canada | 44.63°N, 66.72°W | 18 | 3 | C3, C10, C11 | 0.451 (0.117) | 0.0015 (0.0015) |
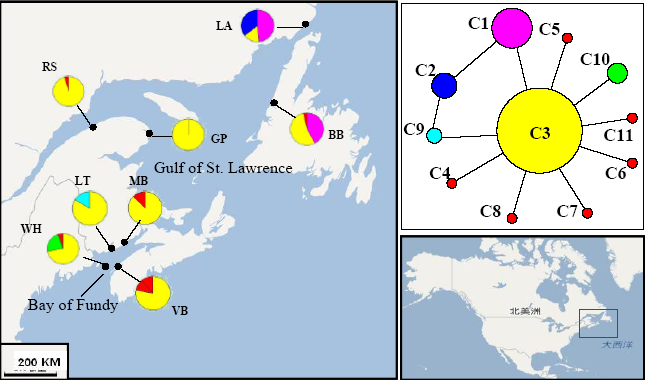
Fig. 2 Distribution of mitochondrial cox2-3 haplotypes of Palmaria palmata populations and median-joining haplotype network. Population codes correspond to those in Table 1.
| 变异来源 Source of variations | 自由度 d.f. | 变异组成 Variance components | 变异百分比 % of variation | 固定指数 Fixation indices |
|---|---|---|---|---|
| 群组间 Among groups | 1 | 0.0409 | 10.12 | FCT = 0.10118NS |
| 种群间 Among populations within groups | 6 | 0.1246 | 34.09 | Fsc = 0.37928*** |
| 种群内 Within populations | 130 | 0.2075 | 55.79 | FST = 0.44509*** |
Table 2 Molecular variance (AMOVA) of Palmaria palmata populations. The eight populations were clustered into two groups: the Gulf of St. Lawrence and the Bay of Fundy.
| 变异来源 Source of variations | 自由度 d.f. | 变异组成 Variance components | 变异百分比 % of variation | 固定指数 Fixation indices |
|---|---|---|---|---|
| 群组间 Among groups | 1 | 0.0409 | 10.12 | FCT = 0.10118NS |
| 种群间 Among populations within groups | 6 | 0.1246 | 34.09 | Fsc = 0.37928*** |
| 种群内 Within populations | 130 | 0.2075 | 55.79 | FST = 0.44509*** |
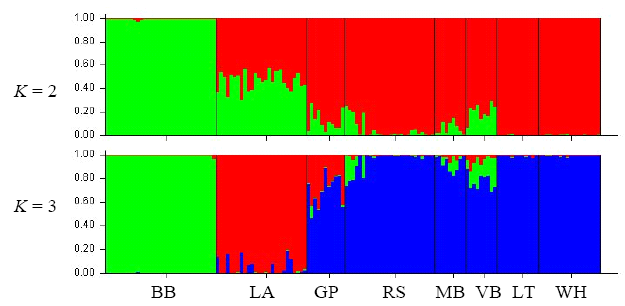
Fig. 3 Population structuring analysis based on RAPD data with STRUCTURE. Each vertical bar indicates the multi-locus genotype of one individual, and colors represent the K virtual clusters. Population codes are the same as Table 1.
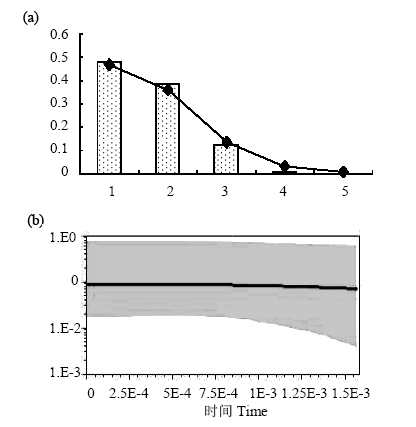
Fig. 4 Demographic history of Palmaria palmata. (a) Mismatch distributions of Palmaria palmata populations. The abscissa indicates the number of pairwise differences between compared sequences. The ordinate is the frequency for each value. Bars represent the observed distribution of pairwise differences, while the solid line shows the expected distribution; (b) Bayesian skyline plots show effective population size as a function of time. The upper and lower limits of grey trend represent the 95% confidence intervals of higher probability density (HPD) analysis.
| 22 | Krebes L, Blank M, Bastrop R (2011) Phylogeography, historical demography and postglacial colonization routes of two amphi-Atlantic distributed amphipods. Systematics and Biodiversity, 9, 259-273. |
| 23 | Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451-1452. |
| 24 | Lindstrom SC, Olsen JL, Stam WT (1996) Recent radiation of the Palmariaceae (rhodophyta). Journal of Phycology, 32, 457-468. |
| 25 | Lindstrom SC, Olsen JL, Stam WT (1997) Postglacial recolonization and the biogeography of Palmaria mollis (Rhodophyta) along the Northeast Pacific coast. Canadian Journal of Botany, 75, 1887-1896. |
| 26 | Lister A, Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B: Biological Sciences, 359, 183-195. |
| 27 | Maggs CA, Castilho R, Foltz D, Henzler C, Jolly MT, Kelly J, Olsen J, Perez KE, Stam W, Väinölä R, Viard F, Wares J (2008) Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology, 89, S108-S122. |
| 28 | Olsen JL, Zechman FW, Hoarau G, Coyer JA, Stam WT, Valero M, Åberg P (2010) The phylogeographic architecture of the fucoid seaweed Ascophyllum nodosum: an intertidal ‘marine tree’ and survivor of more than one glacial- interglacial cycle. Journal of Biogeography, 37, 842-856. |
| 29 | Panova M, Blakeslee AMH, Miller AW, Mäkinen T, Ruiz GM, Johannesson K, André C (2011) Glacial history of the North Atlantic marine snail, Littorina saxatilis, inferred from distribution of mitochondrial DNA lineages. PLoS ONE, 6, e17511. |
| 30 | Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics, 14, 817-818. |
| 31 | Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959. |
| 32 | Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends in Ecology and Evolution, 23, 564-571. |
| 33 | Provan J, Wattier RA, Maggs CA (2005) Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Molecular Ecology, 14, 793-803. |
| 34 | Ray N, Currat M, Excoffier L (2003) Intra-deme molecular diversity in spatially expanding populations. Molecular Biology and Evolution, 20, 76-86. |
| 35 | Riggs SR, Snyder SW, Hine AC, Mearns DL (1996) Hardbottom morphology and relationship to the geologic framework: mid-Atlantic continental shelf. Journal of Sedimentary Research, 66, 830-846. |
| 36 | Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9, 552-569. |
| 37 | Saunders GW (2004) A chronicle of the convoluted systematics of the red algal orders Palmariales and Rhodymeniales (Florideophyceae, Rhodophyta). CEMAR Occasional Notes in Phycology, 1, 1-16. |
| 38 | Strimmer K, Pybus OG (2001) Exploring the demographic history of DNA sequences using the generalized skyline plot. Molecular Biology and Evolution, 18, 2298-2305. |
| 39 | Svendsen JI, Alexanderson H, Astakhov VI, Demidov I, Dowdeswell JA, Funder S, Gataullin V, Henriksen M, Hjort C, Houmark-Nielsen M, Hubberten HW, Ingólfsson O, Jakobsson M, Kjaer KH, Larsen E, Lokrantz H, Lunkka JP, Lyså A, Mangerud J, Matiouchkov A, Murray A, Möller P, Niessen F, Nikolskaya O, Polyak L, Saarnisto M, Siegert C, Siegert MJ, Spielhagen RF, Stein R (2004) Late quaternary ice sheet history of northern Eurasia. Quaternary Science Reviews, 23, 1229-1271. |
| 40 | Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595. |
| 41 | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731-2739. |
| 42 | Teasdale BW, Klein AS (2010) Genetic variation and biogeographical boundaries within the red alga Porphyra umbilicalis (Bangiales, Rhodophyta). Botanica Marina, 53, 417-431. |
| 43 | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. |
| 44 | Vermeij GJ (1991) Anatomy of an invasion: the trans-Arctic interchange. Paleobiology, 17, 281-307. |
| 45 | Victoria M, Miagostovich MP, Ferreira MSR, Vieira CB, Fioretti JM, Leite JPG, Colina R, Cristina J (2009) Bayesian coalescent inference reveals high evolutionary rates and expansion of Norovirus populations. Infection, Genetics and Evolution, 9, 927-932. |
| 46 | Wares JP (2002) Community genetics in the Northwestern Atlantic intertidal. Molecular Ecology, 11, 1131-1144. |
| 47 | Wares JP, Cunningham CW (2001) Phylogeography and historical ecology of the North Atlantic intertidal. Evolution, 55, 2455-2469. |
| 48 | Zuccarello GC, Burger G, West JA, King RJ (1999) A mitochondrial marker for red algal intraspecific relationships. Molecular Ecology, 8, 1443-1447. |
| 49 | Zuccarello GC, West JA (2002) Phylogeography of the Bostrychia calliptera-B. pinnata complex (Rhodomelaceae, Rhodophyta) and divergence rates based on nuclear, mitochondrial and plastid DNA markers. Phycologia, 41, 49-60. |
| 1 | Addison JA, Hart MW (2005) Colonization, dispersal, and hybridization influence phylogeography of North Atlantic sea urchins (Strongylocentrotus droebachiensis). Evolution, 59, 532-543. |
| 2 | Albaina N, Olsen JL, Couceiro L, Ruiz JM, Barreiro R (2012) Recent history of the European Nassarius nitidus (Gastropoda): phylogeographic evidence of glacial refugia and colonization pathways. Marine Biology, 159, 1871-1884. |
| 3 | Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37-48. |
| 4 | Bigg GR, Cunningham CW, Ottersen G, Pogson GH, Wadley MR, Williamson P (2008) Ice-age survival of Atlantic cod: agreement between palaeoecology models and genetics. Proceedings of the Royal Society B: Biological Sciences, 275, 163-172. |
| 5 | Campo D, Molares J, Garcia L, Fernandez-Rueda P, Garcia- Gonzalez C, Garcia-Vazquez E (2010) Phylogeography of the European stalked barnacle (Pollicipes pollicipes): identification of glacial refugia. Marine Biology, 157, 147-156. |
| 6 | Charbit S, Ritz C, Philippon G, Peyaud V, Kageyama M (2007) Numerical reconstructions of the Northern Hemisphere ice sheets through the last glacial-interglacial cycle. Climate of the Past, 3, 15-37. |
| 7 | Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution, 22, 1185-1192. |
| 8 | Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology, 4, 699-710. |
| 9 | Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969-1973. |
| 10 | Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611-2620. |
| 11 | Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Molecular Ecology, 13, 853-864. |
| 12 | Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564-567. |
| 13 | Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131, 479-491. |
| 14 | Flint RF (1940) Pleistocene features of the Atlantic Coastal Plain. American Journal of Science, 238, 757-787. |
| 15 | Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915-925. |
| 16 | Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247-276. |
| 17 | Hoarau G, Coyer JA, Veldsink JH, Stam WT, Olsen JL (2007) Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Molecular Ecology, 16, 3606-3616. |
| 18 | Hu ZM, Guiry MD, Critchley AT, Duan DL (2010) Phylogeographic patterns indicate transatlantic migration from Europe to North America in the red seaweed Chondrus Crispus (Gigartinales, Rhodophyta). Journal of Phycology, 46, 889-900. |
| 19 | Hu ZM, Li W, Li JJ, Duan DL (2011) Post-Pleistocene demographic history of the North Atlantic endemic Irish moss Chondrus crispus: glacial survival, spatial expansion and gene flow. Journal of Evolutionary Biology, 24, 505-517. |
| 20 | Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16, 111-120. |
| 21 | Kingman JFC (1982) On the genealogy of large populations. Journal of Applied Probability, 19, 27-43. |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [3] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [4] | Xianglin Yang, Caiyun Zhao, Junsheng Li, Fangfang Chong, Wenjin Li. Invasive plant species lead to a more clustered community phylogenetic structure: An analysis of herbaceous plants in Guangxi’s national nature reserves [J]. Biodiv Sci, 2024, 32(11): 24175-. |
| [5] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [6] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [7] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [8] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [9] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [10] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [11] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [12] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [13] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [14] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [15] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()