



Biodiv Sci ›› 2011, Vol. 19 ›› Issue (6): 770-778. DOI: 10.3724/SP.J.1003.2011.09149 cstr: 32101.14.SP.J.1003.2011.09149
Special Issue: 中国的海洋生物多样性
Previous Articles Next Articles
Received:2011-08-23
Accepted:2011-12-10
Online:2011-11-20
Published:2011-12-19
Contact:
Jinjun Kan
Supported by:Jinjun Kan, Jun Sun. Bacterial community biodiversity in estuaries and its controlling factors: a case study in Chesapeake Bay[J]. Biodiv Sci, 2011, 19(6): 770-778.
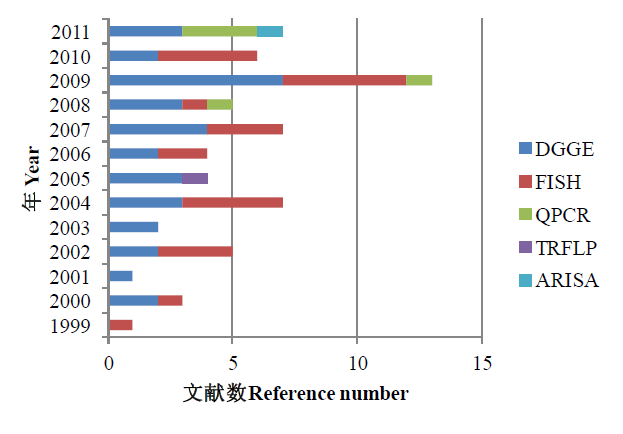
Fig. 1 The molecular techniques for detection diversity of estuary bacteria communities around the world in 1999-2011. DGGE, Denaturing gradient gel electrophoresis; FISH, Fluorescence in situ hybridization; QPCR, Real-time polymerase chain reaction; TRFLP, Terminal restriction fragment length polymorphism; ARISA, Automated ribosomal intergenic spacer analysis. (Searching on November 20th 2011)
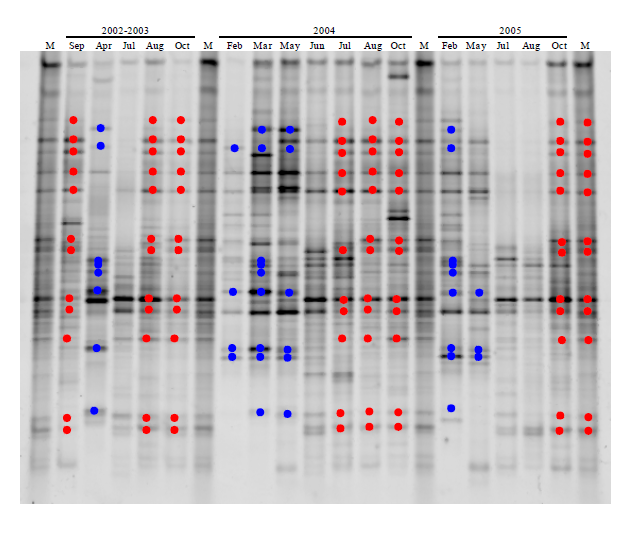
Fig. 4 Fingerprints of Chesapeake Bay bacterial communities during 2002-2005 (middle Bay). Red and blue highlighted the bands appeared in summer and winter, respectively. M, marker.
| [1] |
Amann RI, Krumholz L, Stahl DA (1990) Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. Journal of Bacteriology, 172, 762-770.
URL PMID |
| [2] |
Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF (2000a) Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science, 289, 1902-1906.
URL PMID |
| [3] |
Béjà O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP, Villacorta R, Amjadi M, Garrigues C, Jovanovich SB, Feldman RA, DeLong EF (2000b) Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environmental Microbiology, 2, 516-529.
DOI URL PMID |
| [4] |
Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF (2001) Proteorhodopsin phototrophy in the ocean. Nature, 411, 786-789.
DOI URL PMID |
| [5] | Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2011) GenBank. Nucleic Acids Research, 39(Suppl. 1), D32-D37. |
| [6] | Bouvier TC, del Giorgio PA (2002) Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnology and Oceanography, 47, 453-470. |
| [7] |
Button DK, Schut F, Quang P, Martin R, Robertson BR (1993) Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Applied and Environmental Microbiology, 59, 881-891.
URL PMID |
| [8] |
Cho JC, Giovannoni SJ (2004) Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Applied and Environmental Microbiology, 70, 432-440.
URL PMID |
| [9] |
Cho JC, Vergin KL, Morris RM, Giovannoni SJ (2004) Lentisphaera araneosa gen. nov., sp. nov., a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environmental Microbiology, 6, 611-621.
URL PMID |
| [10] |
Conway T, Schoolnik GK (2003) Microarray expression profiling: capturing a genome-wide protrait of the transcriptome. Molecular Microbiology, 47, 879-889.
URL PMID |
| [11] |
Crump BC, Armbrust EV, Baross JA (1999) Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Applied and Environmental Microbiology, 65, 3192-3204.
URL PMID |
| [12] | Crump BC, Hobbie JE (2005) Synchrony and seasonality in bacterioplankton diversity of two temperate rivers. Limnology and Oceanography, 50, 1718-1729. |
| [13] |
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE (2004) Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Applied and Environmental Microbiology, 70, 1494-1505.
URL PMID |
| [14] | Crump BC, Peterson BJ, Raymond PA, Amon RMW, Rinehart A, McClelland JW, Holmes RM (2009) Circumpolar synchrony in big river bacterioplankton. Proceedings of the National Academy of Sciences, USA, 106, 21208-21212. |
| [15] | Daley RJ, Hobbie JE (1975) Direct counts of aquatic bacteria by a modified epifluorescent technique. Limnology and Oceanography, 20, 875-882. |
| [16] | de la Torre JR, Christianson LM, Béjà O, Suzuki MT, Karl DM, Heidelberg J, DeLong EF (2003) Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proceedings of the National Academy of Sciences,USA, 100, 12830-12835. |
| [17] | del Giorgio PA, Bouvier TC (2002) Linking the physiological and phylogenetic succession in free-living bacterial communities along an estuarine salinity gradient. Limnology and Oceanography, 47, 471-486. |
| [18] |
DeLong EF (2005) Microbial community genomics in the ocean. Nature Reviews Microbiology, 3, 459-469.
DOI URL PMID |
| [19] |
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM (2006) Community genomics among stratified microbial assemblages in the ocean's interior. Science, 311, 496-503.
URL PMID |
| [20] |
Eymann C, Homuth G, Scharf C, Hecker M (2002) Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. Journal of Bacteriology, 184, 2500-2520.
DOI URL PMID |
| [21] | Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S (2006) Annually reoccurring bacterial communities are predictable from ocean conditions. Proceedings of the National Academy of Sciences, USA, 103, 13104-13109. |
| [22] |
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature, 345, 60-63.
URL PMID |
| [23] | Giovannoni SJ, Rappé MS (2000) Evolution, diversity, and molecular ecology of marine prokaryotes. In: Microbial Ecology of the Oceans (ed. Kirchman DL), pp. 47-84. Wiley-Liss, New York. |
| [24] | Glibert PM, Conley DJ, Fisher TR, Harding LWJ, Malone TC (1995) Dynamics of the 1990 winter/spring bloom in Chesapeake Bay. Marine Ecology: Progress Series, 122, 27-43. |
| [25] |
Hahn MW, Lunsdorf H, Wu Q, Schauer M, Höfle MG, Boenigk J, Stadler P (2003) Isolation of novel ultramicrobacterial classified as Actinobacteria from five freshwater habitats in Europe and Asia. Applied and Environmental Microbiology, 69, 1442-1451.
DOI URL PMID |
| [26] |
Heidelberg JF, Heidelberg KB, Colwell RR (2002) Seasonality of Chesapeake Bay bacterioplankton species. Applied and Environmental Microbiology, 68, 5488-5497.
DOI URL PMID |
| [27] | Henriques IS, Alves A, Tacao M, Almeida A, Cunha A, Correia A (2006) Seasonal and spatial variability of free-living bacterial community composition along an estuarine gradient (Ria de Aveiro, Portugal). Estuarine Coastal and Shelf Science, 68, 139-148. |
| [28] | Hollibaugh JT, Wong PS, Murrell MC (2000) Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquatic Microbial Ecology, 21, 103-109. |
| [29] | Hoppe HG (1976) Determination and properties of actively metabolizing heterotrophic bacteria in the sea, investigated by the means of micro-autoradiography. Marine Biology, 36, 291-302. |
| [30] | Jannasch HW, Jones GE (1959) Bacterial populations in sea water as determined by different methods of enumeration. Limnology and Oceanography, 4, 128-139. |
| [31] |
Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M (2002) Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Applied and Environmental Microbiology, 68, 2391-2396.
URL PMID |
| [32] | Kan J, Chen F (2004) Co-monitoring bacterial and dinoflagellates communities by denaturing gradient gel electrophoresis (DGGE) and SSU rDNA sequencing during a dinoflagellate bloom. Acta Oceanologica Sinica, 23, 483-492. |
| [33] | Kan J, Crump BC, Wang K, Chen F (2006a) Bacterioplankton community in Chesapeake Bay: Predictable or random assemblages. Limnology and Oceanography, 51, 2157-2169. |
| [34] | Kan J, Wang K, Chen F (2006b) Temporal variation and detection limit of an estuarine bacterioplankton community analyzed by denaturing gradient gel electrophoresis (DGGE). Aquatic Microbial Ecology, 42, 7-18. |
| [35] |
Kan J, Suzuki M, Wang K, Evans SE, Chen F (2007) High temporal but low spatial heterogeneity of bacterioplankton in the Chesapeake Bay. Applied and Environmental Microbiology, 73, 6776-6789.
DOI URL PMID |
| [36] | Kan J, Evans SE, Chen F, Suzuki MT (2008) Novel estuarine bacterioplankton in rRNA operon libraries from the Chesapeake Bay. Aquatic Microbial Ecology, 51, 55-66. |
| [37] | Kan J, Hanson TE, Chen F (2011) Synchronicity between population structure and proteome profiles: a metaproteomic analysis of Chesapeake Bay bacterial communities. In: Handbook of Molecular Microbial Ecology, Volume I: Metagenomics and Complementary Approaches (ed. de Bruijn FJ), pp. 637-644. Wiley-Blackwell, Hoboken. |
| [38] |
Kan J, Hanson TE, Ginter JM, Wang K, Chen F (2005) Metaproteomic analysis of Chesapeake Bay microbial communities. Saline Systems, 1, 7.
DOI URL PMID |
| [39] |
Kent AD, Jones SE, Yannarell AC, Graham JM, Lauster GH, Kratz TK, Triplett EW (2004) Annual patterns in bacterioplankton community variability in a humic lake. Microbial Ecology, 48, 550-560.
DOI URL PMID |
| [40] |
Kirchman DL, Rich JH (1997) Regulation of bacterial growth rates by dissolved organic carbon and temperature in the equatorial Pacific Ocean. Microbial Ecology, 33, 11-20.
DOI URL PMID |
| [41] | Lopez MF (1999) Proteome analysis I. Gene products are where the biological action is. Journal of Chromatography B, 722, 191-202. |
| [42] | Malone TC, Kemp WM, Ducklow HW, Boynton WR, Tuttle JH, Jonas RB (1986) Lateral variation in the production and fate of phytoplankton in a partially stratified estuary. Marine Ecology-Progress Series, 32, 149-160. |
| [43] |
Martinez A, Tyson GW, DeLong EF (2009) Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environmental Microbiology, 12, 222-238.
DOI URL PMID |
| [44] | Monsen NE, Cloern JE, Lucas LV, Monismith SG (2002) A comment on the use of flushing time, residence time, and age as transport time scales. Limnology and Oceanography, 47, 1545-1553. |
| [45] |
Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K, DeLong EF (1998) Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Applied and Environmental Microbiology, 64, 2585-2595.
DOI URL PMID |
| [46] | Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, Rinta-Kanto JM, Sharma S, Sun S, Varaljay V, Vila-Costa M, Westrich JR, Moran MA (2010) Genome characteristics of a generalist marine bacterial lineage. ISME Journal, 4, 784-798. |
| [47] | Nixon SW, Ammerman JW, Atkinson LP, Berounsky VM, Billen G, Boicourt WC, Boynton WR, Church TM, Ditoro DM, Elmgren R, Garber JH, Giblin AE, Jahnke RA, Owens NJP, Pilson MEQ, Seitzinger SP (1996) The fate of nitrogen and phosphorus at the land sea margin of the North Atlantic Ocean. Biogeochemistry, 35, 141-180. |
| [48] |
Olsen GJ, Lane DJ, Giovannoni J, Pace NR (1986) Microbial ecology and evolution: a ribosomal RNA approach. Annual Review of Microbiology, 40, 337-365.
URL PMID |
| [49] | Pace NR, Stahl DA, Olsen GJ, Lane DJ (1985) Analyzing natural microbial populations by rRNA sequences. American Society of Microbiology News, 51, 4-12. |
| [50] |
Pernthaler J, Glöckner FO, Unterholzner S, Alfreider A, Psenner R, Amann R (1998) Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Applied and Environmental Microbiology, 64, 4299-4306.
DOI URL PMID |
| [51] |
Petersohn A, Brigulla M, Haas S, Hoheisel JD, Volker U, Hecker M (2001) Global analysis of the general stress response of Bacillus subtilis. Journal of Bacteriology, 183, 5617-5631.
URL PMID |
| [52] |
Pinhassi M, Sala M, Havskum H, Peters F, Guadayol O, Malits A, Marrase C (2004) Changes in bacterioplankton composition under different phytoplankton regimens. Applied and Environmental Microbiology, 70, 6753-6766.
URL PMID |
| [53] | Pomeroy LR, Wiebe WJ (2001) Temperature and substrates as interactive limiting factors for marine heterotrophic bacteria. Aquatic Microbial Ecology, 23, 187-204. |
| [54] | Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnology and Oceanography, 25, 943-948. |
| [55] |
Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake II RC, Shah M, Hettich RL, Banfield JF (2005) Community proteomics of a natural microbial biofilm. Science, 308, 1915-1920.
DOI URL PMID |
| [56] |
Rappé MS, Cannon SA, Vergin KL, Giovannoni SJ (2002) Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature, 418, 630-633.
URL PMID |
| [57] |
Riemann L, Steward GF, Azam F (2000) Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Applied and Environmental Microbiology, 66, 578-587.
DOI URL PMID |
| [58] |
Riesenfeld CS, Schloss PD, Handelsman J (2004) Metagenomics: genomic analysis of microbial communities. Annual Review of Genetics, 38, 525-552.
DOI URL PMID |
| [59] | SAS Institute Inc (1992) SAS User's Guide. SAS Institute Inc. |
| [60] |
Sekiguchi H, Koshikawa H, Hiroki M, Murakami S, Xu K, Watanabe M, Nakahara T, Zhu M, Uchiyama H (2002) Bacterial distribution and phylogenetic diversity in the Changjiang estuary before the construction of the Three Gorges Dam. Microbial Ecology, 43, 82-91.
URL PMID |
| [61] | Selje N, Simon M (2003) Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquatic Microbial Ecology, 30, 221-237. |
| [62] | Shiah F-K, Ducklow HW (1994) Temperature regulation of heterotrophic bacterioplankton abundance, production, and specific growth rate. Limnology and Oceanography, 39, 1243-1258. |
| [63] |
Simon C, Daniel R (2011) Metagenomic analyses: past and future trends. Applied and Environmental Microbiology, 77, 1153-1161.
URL PMID |
| [64] | Smith EM, Kemp WM (2003) Planktonic and bacterial respiration along an estuarine gradient: response to carbon and nutrient enrichment. Aquatic Microbial Ecology, 30, 251-261. |
| [65] | Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, Carlson CA, Smith RD, Giovanonni SJ (2008) Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME Journal, 3, 93-105. |
| [66] |
Staley JT, Konopka A (1985) Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annual Review of Microbiology, 39, 321-346.
URL PMID |
| [67] |
Stevenson, BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA (2004) New strategies for cultivation and detection of previously uncultured microbes. Applied and Environmental Microbiology, 70, 4748-4755.
URL PMID |
| [68] | Sun J, Feng YY, Zhang YH, Hutchins DA (2007) Fast microzooplankton grazing on fast-growing, low-biomass phytoplankton ― A case study in spring in the Chesapeake Bay, Delaware Inland Bays and the Delaware Bay. Hydrobiology, 589, 127-139. |
| [69] |
Temperton B, Gilbert JA, Quinn JP, McGrath JW (2011) Novel analysis of oceanic surface water metagenomes suggests importance of polyphosphate metabolism in oligotrophic environments. PLoS ONE, 6, e16499. doi: 10.1371/journal. pone.0016499
URL PMID |
| [70] |
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM (2005) Comparative metagenomics of microbial communities. Science, 308, 554-557.
DOI URL PMID |
| [71] | Troussellier M, Schäfer H, Batailler N, Bernard L, Courties C, Lebaron P, Muyzer G, Servais P, Vives-Rego J (2002) Bacterial activity and genetic richness along an estuarine gradient (Rhone River plume, France). Aquatic Microbial Ecology, 28, 13-24. |
| [72] |
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature, 428, 37-43.
DOI URL PMID |
| [73] |
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science, 304, 66-74.
URL PMID |
| [74] | Wikner J, Hagström A (1991) Annual study of bacteri- oplankton community dynamics. Limnology and Oceanography, 36, 1313-1324. |
| [75] |
Wilmes P, Bond PL (2004) The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environmental Microbiology, 6, 911-920.
DOI URL PMID |
| [76] |
ZoBell CE (1946) Marine Microbiology. Chronica Botanica, Waltham, Mass.
DOI URL PMID |
| [1] | Xiaoqiang Lu, Shanshan Dong, Yue Ma, Xu Xu, Feng Qiu, Mingyue Zang, Yaqiong Wan, Luanxin Li, Cigang Yu, Yan Liu. Current status, challenges, and prospects of frontier technologies in biodiversity conservation applications [J]. Biodiv Sci, 2025, 33(4): 24440-. |
| [2] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [3] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [4] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [5] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [6] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [7] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [8] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [9] | Zhaoyang Jing, Keguang Cheng, Heng Shu, Yongpeng Ma, Pingli Liu. Whole genome resequencing approach for conservation biology of endangered plants [J]. Biodiv Sci, 2023, 31(5): 22679-. |
| [10] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [11] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [12] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [13] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [14] | Congcong Lu, Qian Liu, Xiaolei Huang. Mitochondrial genome data of three aphid species [J]. Biodiv Sci, 2022, 30(7): 22204-. |
| [15] | Ruiliang Zhu, Xiaoying Ma, Chang Cao, Ziyin Cao. Advances in research on bryophyte diversity in China [J]. Biodiv Sci, 2022, 30(7): 22378-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()
