



生物多样性 ›› 2022, Vol. 30 ›› Issue (4): 21457. DOI: 10.17520/biods.2021457 cstr: 32101.14.biods.2021457
收稿日期:2021-11-15
接受日期:2022-02-10
出版日期:2022-04-20
发布日期:2022-02-28
通讯作者:
毛晓伟,赵毅强
作者简介:maoxiaowei@ivpp.ac.cn基金资助:
Xinyu Cai1, Xiaowei Mao2,*( ), Yiqiang Zhao1,*(
), Yiqiang Zhao1,*( )
)
Received:2021-11-15
Accepted:2022-02-10
Online:2022-04-20
Published:2022-02-28
Contact:
Xiaowei Mao,Yiqiang Zhao
摘要:
驯化动物伴随了人类上万年的进化历程。在驯化过程中, 家养动物表型和行为特征发生巨大改变, 并与人类生产生活的进步相适应。研究动物驯化问题对于了解遗传多样性及适应性进化, 解析复杂性状的遗传机制等具有重要的意义, 成为生物学领域的重点研究内容之一。本文专注于动物驯化的初始阶段, 首先介绍了驯化起源的时间与地点、驯化途径和驱动因、驯化后的扩散和品种选育; 其次从驯化对象的角度着重介绍了考古学、分子和群体遗传学两方面的家养动物驯化起源的研究策略、优势与不足, 以及未来的发展方向; 最后, 我们综合多方面的证据, 介绍了在中国驯化的家猪(Sus domesticus)和家鸡(Gallus gallus domesticus)以及其他主要家养动物考古和分子水平上驯化起源的研究进展。本文整合多种证据为家养动物驯化起源的研究提供了相对完整的视角和新的思路。
蔡新宇, 毛晓伟, 赵毅强 (2022) 家养动物驯化起源的研究方法与进展. 生物多样性, 30, 21457. DOI: 10.17520/biods.2021457.
Xinyu Cai, Xiaowei Mao, Yiqiang Zhao (2022) Methods and research progress on the origin of animal domestication. Biodiversity Science, 30, 21457. DOI: 10.17520/biods.2021457.
| 学名 Scientific names | 地理起源 Geographic origin | 野生祖先 Wild ancestor | 前驯化时间(距今年限) Pre-domestication time (BP) | 形态学转变(距今年限) Time of morphological changes (BP) | 驯化途径 Pathway to domestication | ||
|---|---|---|---|---|---|---|---|
| 起始 Start | 完成 Finish | 起始 Start | 完成 Finish | ||||
| 山羊 Capra hircus | 西南亚 Southwest Asia | 野山羊 C. aegagrus | 10,500 | 9,750 | 9,750 | 8,000 | 猎食 Prey |
| 绵羊 Ovis aries | 西南亚 Southwest Asia | 亚洲盘羊 O. orientalis | 10,500 | 9,750 | 9,750 | 8,000 | 猎食 Prey |
| 普通牛 Bos taurus | 西南亚 Southwest Asia | 原牛欧亚亚种 B. p. primigenius | 10,500 | 10,250 | 10,250 | 8,000 | 猎食 Prey |
| 瘤牛 Bos indicus | 南亚 South Asia | 原牛印度亚种 B. p. mauretanicus | × | × | 8,000 | 6,500 | 猎食 Prey |
| 猪 Sus domesticus | 西南亚 Southwest Asia | 欧亚野猪 S. scrofa | 11,500 | 9,750 | 10,250 | 9,000 | 共生 Commensal |
| 东亚 East Asia | 欧亚野猪 S. scrofa | × | × | 8,500 | 6,000 | 共生 Commensal | |
| 美洲驼 Lama glama | 南美洲 South America | 原驼 L. guanicoe | × | × | 6,000 | 4,000 | 猎食 Prey |
| 羊驼 Vicugna pacos | 南美洲 South America | 原驼 L. guanicoe | × | × | 5,000 | 3,000 | 猎食 Prey |
| 马 Equus caballus | 中亚 Central Asia | 野马 E. ferus | 6,750 | 5,500 | 5,500 | 4,000 | 定向 Directed |
| 驴 Equus asinus | 北非 North Africa | 非洲野驴 E. africanus | × | × | 5,500 | 3,500 | 定向 Directed |
| 水牛 Bubalus bubalis | 南亚 South Asia | 野水牛 B. arnee | × | × | 4,500 | × | 猎食 Prey |
| 双峰驼 Camelus bactrianus | 中亚 Central Asia | × | × | × | 4,500 | × | 定向 Directed |
| 单峰驼 Camelus dromedarius | 阿拉伯半岛 Arabia | × | × | × | 3,000 | × | 定向 Directed |
| 鸡 Gallus gallus domesticus | 东亚/东南亚 East Asia/Southeast Asia | 红原鸡 G. gallus | × | × | 4,000 | × | 共生 Commensal |
| 家鸭 Anas platyrhynchos domesticus | 东亚/东南亚 East Asia/Southeast Asia | 绿头鸭 A. platyrhynchos | × | × | 1,000 | × | 共生 Commensal |
表1 主要家养动物的驯化起源信息。根据Larson和Fuller (2014)、Larson等(2014)和Teletchea (2019)整理。内容包括地理起源、野生祖先、时间起源(前驯化时期和形态学改变时期)和驯化途径。其中前驯化时期是在发生骨骼形态学和性状变化之前出现的特殊集约化管理现象, 是迈向驯化时必不可少的探索阶段。“×”表示此处目前尚未取得可靠证据或者存在较大争议。
Table1 The summary of the origin of domestication of major domestic animals, collected from Larson & Fuller (2014), Larson (2014), and Teletchea (2019). This table includes geographic origin, wild ancestor, domestication time (pre-domestication and the period of morphological changes), and domestication pathways. Among them, the pre-domestication period is a specific intensive management that occurs before the changes in bone morphology and other traits, and it is an indispensable preparation before domestication. “×” means there is no reliable evidence or it is still under debate.
| 学名 Scientific names | 地理起源 Geographic origin | 野生祖先 Wild ancestor | 前驯化时间(距今年限) Pre-domestication time (BP) | 形态学转变(距今年限) Time of morphological changes (BP) | 驯化途径 Pathway to domestication | ||
|---|---|---|---|---|---|---|---|
| 起始 Start | 完成 Finish | 起始 Start | 完成 Finish | ||||
| 山羊 Capra hircus | 西南亚 Southwest Asia | 野山羊 C. aegagrus | 10,500 | 9,750 | 9,750 | 8,000 | 猎食 Prey |
| 绵羊 Ovis aries | 西南亚 Southwest Asia | 亚洲盘羊 O. orientalis | 10,500 | 9,750 | 9,750 | 8,000 | 猎食 Prey |
| 普通牛 Bos taurus | 西南亚 Southwest Asia | 原牛欧亚亚种 B. p. primigenius | 10,500 | 10,250 | 10,250 | 8,000 | 猎食 Prey |
| 瘤牛 Bos indicus | 南亚 South Asia | 原牛印度亚种 B. p. mauretanicus | × | × | 8,000 | 6,500 | 猎食 Prey |
| 猪 Sus domesticus | 西南亚 Southwest Asia | 欧亚野猪 S. scrofa | 11,500 | 9,750 | 10,250 | 9,000 | 共生 Commensal |
| 东亚 East Asia | 欧亚野猪 S. scrofa | × | × | 8,500 | 6,000 | 共生 Commensal | |
| 美洲驼 Lama glama | 南美洲 South America | 原驼 L. guanicoe | × | × | 6,000 | 4,000 | 猎食 Prey |
| 羊驼 Vicugna pacos | 南美洲 South America | 原驼 L. guanicoe | × | × | 5,000 | 3,000 | 猎食 Prey |
| 马 Equus caballus | 中亚 Central Asia | 野马 E. ferus | 6,750 | 5,500 | 5,500 | 4,000 | 定向 Directed |
| 驴 Equus asinus | 北非 North Africa | 非洲野驴 E. africanus | × | × | 5,500 | 3,500 | 定向 Directed |
| 水牛 Bubalus bubalis | 南亚 South Asia | 野水牛 B. arnee | × | × | 4,500 | × | 猎食 Prey |
| 双峰驼 Camelus bactrianus | 中亚 Central Asia | × | × | × | 4,500 | × | 定向 Directed |
| 单峰驼 Camelus dromedarius | 阿拉伯半岛 Arabia | × | × | × | 3,000 | × | 定向 Directed |
| 鸡 Gallus gallus domesticus | 东亚/东南亚 East Asia/Southeast Asia | 红原鸡 G. gallus | × | × | 4,000 | × | 共生 Commensal |
| 家鸭 Anas platyrhynchos domesticus | 东亚/东南亚 East Asia/Southeast Asia | 绿头鸭 A. platyrhynchos | × | × | 1,000 | × | 共生 Commensal |
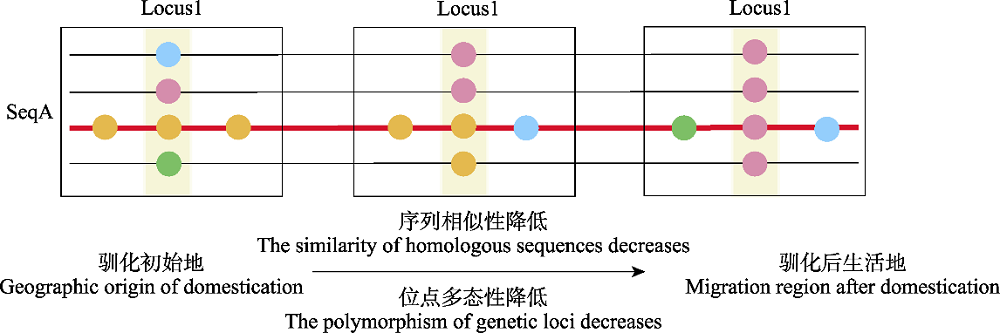
图1 基于分子手段判断驯化起源的策略示意图。不同颜色的圆圈代表不同的基因型。纵向为基因座, 指示位点多态性; 横向将基因座连成序列, 指示序列相似性。伴随驯化的过程, 动物从驯化初始地迁移到各地, 同源序列的相似性降低, 迁移样本在同源位点上的多态性降低。
Fig. 1 Strategies for inferring the origin of domestication based on molecular evidences. Circles with different colors represent different genotypes. The vertical direction represents allelic polymorphism of each genetic locus; the horizontal sequence represents the nucleotide sequence. During the process of domestication, animals migrate from domestication site to other places, with decreased similarity in nucleotide sequences and decreased polymorphism of genetic loci.
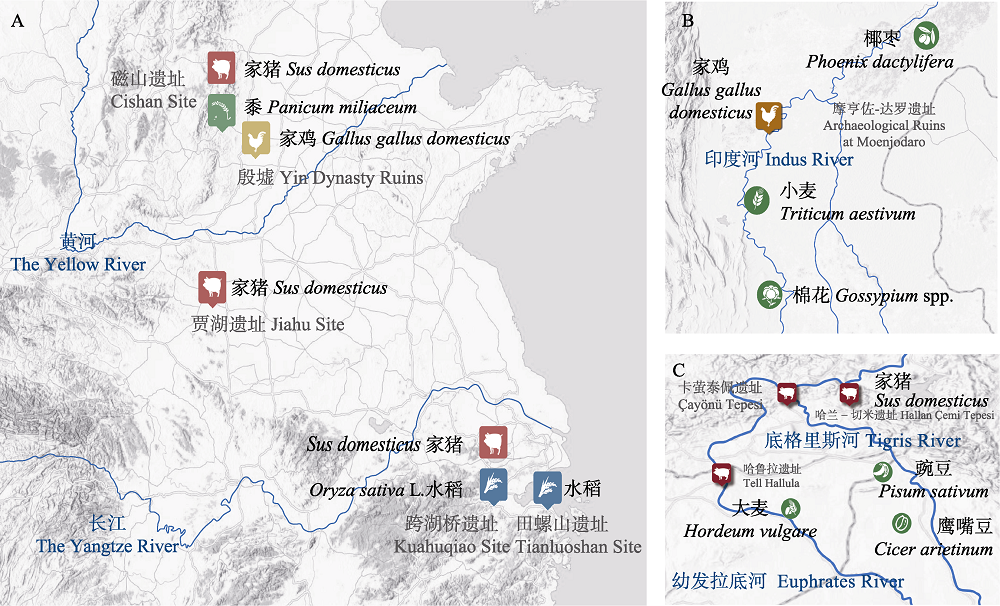
图2 家猪与家鸡早期考古遗址及周边河流、农作物驯化情况。(A)中国长江黄河孕育的中华文明。(B)印度河孕育的印度河文明。(C)新月沃土孕育的美索不达米亚文明。
Fig. 2 Early archaeological sites of domesticated pigs and chickens, with surrounding rivers and crops. (A) Chinese civilization nurtured by the Yangtze River and Yellow River. (B) The Indus civilization nurtured by the Indus. (C) Mesopotamian civilization nurtured by the Fertile Crescent.
| [1] |
Ai H, Fang X, Yang B, Huang Z, Chen H, Mao L, Zhang F, Zhang L, Cui L, He W, Yang J, Yao X, Zhou L, Han L, Li J, Sun S, Xie X, Lai B, Su Y, Lu Y, Yang H, Huang T, Deng W, Nielsen R, Ren J, Huang L (2015) Adaptation and possible ancient inter species introgression in pigs identified by whole-genome sequencing. Nature Genetics, 47, 217- 225.
DOI URL |
| [2] |
Alberto FJ, Boyer F, Orozco-terWengel P, Streeter I, Servin B, de Villemereuil P, Benjelloun B, Librado P, Biscarini F, Colli L, Barbato M, Zamani W, Alberti A, Engelen S, Stella A, Joost S, Ajmone-Marsan P, Negrini R, Orlando L, Rezaei HR, Naderi S, Clarke L, Flicek P, Wincker P, Coissac E, Kijas J, Tosser-Klopp G, Chikhi A, Bruford MW, Taberlet P, Pompanon F (2018) Convergent genomic signatures of domestication in sheep and goats. Nature Communications, 9, 813.
DOI URL |
| [3] |
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655-1664.
DOI PMID |
| [4] | Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, Hale ML, Campos PF, Samaniego JA, Gilbert MTP, Willerslev E, Zhang G, Scofield RP, Holdaway RN, Bunce M (2012) The half-life of DNA in bone:Measuring decay kinetics in 158 dated fossils. Proceedings: Biological Sciences, 279, 4724-4733. |
| [5] | Arbuckle BS (2005) Experimental animal domestication and its application to the study of animal exploitation in prehistory. In:Proceedings of the 19th Conference of the International Council of Archaeozoology, pp. 18-33, Durham. |
| [6] | Bosse M (2018) A genomics perspective on pig domestication. In: Animal Domestication (ed. Teletchea F). IntechOpen, London. |
| [7] |
Bosse M, Megens HJ, Derks MFL, de Cara ÁMR, Groenen MAM (2019) Deleterious alleles in the context of domestication, inbreeding, and selection. Evolutionary Applications, 12, 6-17.
DOI URL |
| [8] | Boyko AR, Boyko RH, Boyko CM, Parker HG, Castelhano M, Corey L, Degenhardt JD, Auton A, Hedimbi M, Kityo R, Ostrander EA, Schoenebeck J, Todhunter RJ, Jones P, Bustamante CD (2009) Complex population structure in African village dogs and its implications for inferring dog domestication history. Proceedings of the National Academy of Sciences, USA, 106, 13903-13908. |
| [9] |
Brown SK, Pedersen NC, Jafarishorijeh S, Bannasch DL, Ahrens KD, Wu JT, Okon M, Sacks BN (2011) Phylogenetic distinctiveness of Middle Eastern and Southeast Asian village dog Y chromosomes illuminates dog origins. PLoS ONE, 6, e28496.
DOI URL |
| [10] |
Brown TA, Jones MK, Powell W, Allaby RG (2009) The complex origins of domesticated crops in the Fertile Crescent. Trends in Ecology & Evolution, 24, 103-109.
DOI URL |
| [11] |
Browning SR, Browning BL (2012) Identity by descent between distant relatives: Detection and applications. Annual Review of Genetics, 46, 617-633.
DOI PMID |
| [12] | Cai DW, Zhang NF, Zhao X (2021) Research on the origins and spread of Chinese goat. Cultural Relics in Southern China, (1), 191-200. (in Chinese with English abstract) |
| [ 蔡大伟, 张乃凡, 赵欣 (2021) 中国山羊的起源与扩散研究. 南方文物, (1), 191-200.] | |
| [13] | Chen NB (2019) Whole-Genome Resequencing Reveals World-Wide Ancestry and Adaptive Introgression Events of Domesticated Cattle in East Asia. PhD dissertation, Northwest A&F University, Yangling. (in Chinese with English abstract) |
| [ 陈宁博 (2019) 全基因组重测序分析揭示东亚家牛的祖先与多重适应性基因渗入. 博士学位论文, 西北农林科技大学, 杨凌.] | |
| [14] |
Chen SY, Lin BZ, Baig M, Mitra B, Lopes RJ, Santos AM, Magee DA, Azevedo M, Tarroso P, Sasazaki S, Ostrowski S, Mahgoub O, Chaudhuri TK, Zhang YP, Costa V, Royo LJ, Goyache F, Luikart G, Boivin N, Fuller DQ, Mannen H, Bradley DG, Beja-Pereira A (2010) Zebu cattle are an exclusive legacy of the south Asia Neolithic. Molecular Biology and Evolution, 27, 1-6.
DOI URL |
| [15] | Chen SY, Zhang YP (2006) Genetic methods and applications in the study of the origin of domestic animals. Chinese Science Bulletin, 51, 2469-2475. (in Chinese) |
| [ 陈善元, 张亚平 (2006) 家养动物起源研究的遗传学方法及其应用. 科学通报, 51, 2469-2475.] | |
| [16] |
Chessa B, Pereira F, Arnaud F, Amorim A, Goyache F, Mainland I, Kao RR, Pemberton JM, Beraldi D, Stear MJ, Alberti A, Pittau M, Iannuzzi L, Banabazi MH, Kazwala RR, Zhang YP, Arranz JJ, Ali BA, Wang ZL, Uzun M, Dione MM, Olsaker I, Holm LE, Saarma U, Ahmad S, Marzanov N, Eythorsdottir E, Holland MJ, Ajmone-Marsan P, Bruford MW, Kantanen J, Spencer TE, Palmarini M (2009) Revealing the history of sheep domestication using retrovirus integrations. Science, 324, 532-536.
DOI URL |
| [17] |
Cucchi T, Hulme-Beaman A, Yuan J, Dobney K (2011) Early Neolithic pig domestication at Jiahu, Henan Province, China: Clues from molar shape analyses using geometric morphometric approaches. Journal of Archaeological Science, 38, 11-22.
DOI URL |
| [18] | Currat M, Ruedi M, Petit RJ, Excoffier L (2008) The hidden side of invasions: Massive introgression by local genes. Evolution, 62, 1908-1920. |
| [19] |
Daly KG, Maisano DP, Mullin VE, Scheu A, Mattiangeli V, Teasdale MD, Hare AJ, Burger J, Verdugo MP, Collins MJ, Kehati R, Erek CM, Bar-Oz G, Pompanon F, Cumer T, Çakırlar C, Mohaseb AF, Decruyenaere D, Davoudi H, Çevik Ö, Rollefson G, Vigne JD, Khazaeli R, Fathi H, Doost SB, Rahimi SR, Vahdati AA, Sauer EW, Azizi KH, Maziar S, Gasparian B, Pinhasi R, Martin L, Orton D, Arbuckle BS, Benecke N, Manica A, Horwitz LK, Mashkour M, Bradley DG (2018) Ancient goat genomes reveal mosaic domestication in the Fertile Crescent. Science, 361, 85-88.
DOI URL |
| [20] | Der Sarkissian C, Allentoft ME, Ávila-Arcos MC, Barnett R, Campos PF, Cappellini E, Ermini L, Fernández R, da Fonseca R, Ginolhac A, Hansen AJ, Jónsson H, Korneliussen T, Margaryan A, Martin MD, Moreno-Mayar JV, Raghavan M, Rasmussen M, Velasco MS, Schroeder H, Schubert M, Seguin-Orlando A, Wales N, Gilbert MTP, Willerslev E, Orlando L (2015) Ancient genomics. Philosophical Transactions of the Royal Society B: Biological Sciences, 370, 20130387. |
| [21] | Diamond J (1997) Guns, Germs, and Steel: The Fates of Human Societies. W. W. Norton, New York. |
| [22] |
Diamond J (2002) Evolution, consequences and future of plant and animal domestication. Nature, 418, 700-707.
DOI URL |
| [23] |
Diamond J, Bellwood P (2003) Farmers and their languages: The first expansions. Science, 300, 597-603.
PMID |
| [24] | Driscoll CA, MacDonald DW, O’Brien SJ (2009) From wild animals to domestic pets, an evolutionary view of domestication. Proceedings of the National Academy of Sciences, USA, 106, 9971-9978. |
| [25] |
Eriksson J, Larson G, Gunnarsson U, Bed'hom B, Tixier- Boichard M, Strömstedt L, Wright D, Jungerius A, Vereijken A, Randi E, Jensen P, Andersson L (2008) Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genetics, 4, e1000010.
DOI URL |
| [26] |
Ervynck A, Dobney K, Hongo H, Meadow R (2001) Born free? New evidence for the status of Sus scrofa at Neolithic Cayonu Tepesi. Paléorient, 27, 47-73.
DOI URL |
| [27] |
Felius M, Beerling ML, Buchanan D, Theunissen B, Koolmees P, Lenstra J (2014) On the history of cattle genetic resources. Diversity, 6, 705-750.
DOI URL |
| [28] |
Frantz L, Meijaard E, Gongora J, Haile J, Groenen MAM, Larson G (2016) The evolution of Suidae. Annual Review of Animal Biosciences, 4, 61-85.
DOI URL |
| [29] |
Frantz LAF, Bradley DG, Larson G, Orlando L (2020) Animal domestication in the era of ancient genomics. Nature Reviews Genetics, 21, 449-460.
DOI URL |
| [30] | Frantz LAF, Haile J, Lin AT, Scheu A, Geörg C, Benecke N, Alexander M, Linderholm A, Mullin VE, Daly KG, Battista VM, Price M, Gron KJ, Alexandri P, Arbogast RM, Arbuckle B, Bӑlӑşescu A, Barnett R, Bartosiewicz L, Baryshnikov G, Bonsall C, Borić dušan, Boroneanţ A, Bulatović J, Çakirlar C, Carretero JM, Chapman J, Church M, Crooijmans R, de Cupere B, Detry C, Dimitrijevic V, Dumitraşcu V, du Plessis L, Edwards CJ, Erek CM, Erim-Özdoğan A, Ervynck A, Fulgione D, Gligor M, Götherström A, Gourichon L, Groenen MAM, Helmer D, Hongo H, Horwitz LK, Irving-Pease EK, Lebrasseur O, Lesur J, Malone C, Manaseryan N, Marciniak A, Martlew H, Mashkour M, Matthews R, Matuzeviciute GM, Maziar S, Meijaard E, McGovern T, Megens HJ, Miller R, Mohaseb AF, Orschiedt J, Orton D, Papathanasiou A, Pearson MP, Pinhasi R, Radmanović D, Ricaut FX, Richards M, Sabin R, Sarti L, Schier W, Sheikhi S, Stephan E, Stewart JR, Stoddart S, Tagliacozzo A, Tasić N, Trantalidou K, Tresset A, Valdiosera C, van den Hurk Y, van Poucke S, Vigne JD, Yanevich A, Zeeb-Lanz A, Triantafyllidis A, Gilbert MTP, Schibler J, Rowley-Conwy P, Zeder M, Peters J, Cucchi T, Bradley DG, Dobney K, Burger J, Evin A, Girdland-Flink L, Larson G (2019) Ancient pigs reveal a near-complete genomic turnover following their introduction to Europe. Proceedings of the National Academy of Sciences, USA, 116, 17231-17238. |
| [31] |
Frantz LAF, Schraiber JG, Madsen O, Megens HJ, Cagan A, Bosse M, Paudel Y, Crooijmans RPMA, Larson G, Groenen MAM (2015) Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nature Genetics, 47, 1141-1148.
DOI |
| [32] | Fu QM, Meyer M, Gao X, Stenzel U, Burbano HA, Kelso J, Pääbo S (2013) DNA analysis of an early modern human from Tianyuan Cave, China. Proceedings of the National Academy of Sciences, USA, 110, 2223-2227. |
| [33] | Fumihito A, Miyake T, Sumi S, Takada M, Ohno S, Kondo N (1994) One subspecies of the red junglefowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proceedings of the National Academy of Sciences, USA, 91, 12505-12509. |
| [34] | Fumihito A, Miyake T, Takada M, Shingu R, Endo T, Gojobori T, Kondo N, Ohno S (1996) Monophyletic origin and unique dispersal patterns of domestic fowls. Proceedings of the National Academy of Sciences, USA, 93, 6792-6795. |
| [35] |
Gansauge MT, Meyer M (2013) Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nature Protocols, 8, 737-748.
DOI URL |
| [36] |
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: Ten years of next-generation sequencing technologies. Nature Reviews Genetics, 17, 333-351.
DOI PMID |
| [37] |
Groenen MAM, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, Li S, Larkin DM, Kim H, Frantz LAF, Caccamo M, Ahn H, Aken BL, Anselmo A, Anthon C, Auvil L, Badaoui B, Beattie CW, Bendixen C, Berman D, Blecha F, Blomberg J, Bolund L, Bosse M, Botti S, Bujie Z, Bystrom M, Capitanu B, Carvalho-Silva D, Chardon P, Chen C, Cheng R, Choi SH, Chow W, Clark RC, Clee C, Crooijmans RPMA, Dawson HD, Dehais P, de Sapio F, Dibbits B, Drou N, Du ZQ, Eversole K, Fadista J, Fairley S, Faraut T, Faulkner GJ, Fowler KE, Fredholm M, Fritz E, Gilbert JGR, Giuffra E, Gorodkin J, Griffin DK, Harrow JL, Hayward A, Howe K, Hu ZL, Humphray SJ, Hunt T, Hornshøj H, Jeon JT, Jern P, Jones M, Jurka J, Kanamori H, Kapetanovic R, Kim J, Kim JH, Kim KW, Kim TH, Larson G, Lee K, Lee KT, Leggett R, Lewin HA, Li Y, Liu W, Loveland JE, Lu Y, Lunney JK, Ma J, Madsen O, Mann K, Matthews L, McLaren S, Morozumi T, Murtaugh MP, Narayan J, Truong Nguyen D, Ni P, Oh SJ, Onteru S, Panitz F, Park EW, Park HS, Pascal G, Paudel Y, Perez-Enciso M, Ramirez-Gonzalez R, Reecy JM, Rodriguez-Zas S, Rohrer GA, Rund L, Sang Y, Schachtschneider K, Schraiber JG, Schwartz J, Scobie L, Scott C, Searle S, Servin B, Southey BR, Sperber G, Stadler P, Sweedler JV, Tafer H, Thomsen B, Wali R, Wang J, Wang J, White S, Xu X, Yerle M, Zhang G, Zhang J, Zhang J, Zhao S, Rogers J, Churcher C, Schook LB (2012) Analyses of pig genomes provide insight into porcine demography and evolution. Nature, 491, 393- 398.
DOI URL |
| [38] |
Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, Pilling D, Negrini R, Finlay EK, Jianlin H, Groeneveld E, Weigend S, Consortium G (2010) Genetic diversity in farm animals-A review. Animal Genetics, 41, 6-31.
DOI URL |
| [39] |
Guatelli-Steinberg D, Ferrell RJ, Spence J (2012) Linear enamel hypoplasia as an indicator of physiological stress in great apes: Reviewing the evidence in light of enamel growth variation. American Journal of Physical Anthropology, 148, 191-204.
DOI PMID |
| [40] |
Guatelli-Steinberg D, Larsen CS, Hutchinson DL (2004) Prevalence and the duration of linear enamel hypoplasia: A comparative study of Neandertals and Inuit foragers. Journal of Human Evolution, 47, 65-84.
PMID |
| [41] |
Guiry EJ, Grimes V (2013) Domestic dog (Canis familiaris) diets among coastal Late Archaic groups of northeastern North America: A case study for the canine surrogacy approach. Journal of Anthropological Archaeology, 32, 732-745.
DOI URL |
| [42] |
Guo Y, Lillie M, Zan Y, Beranger J, Martin A, Honaker CF, Siegel PB, Carlborg Ö (2019) A genomic inference of the white Plymouth rock genealogy. Poultry Science, 98, 5272- 5280.
DOI PMID |
| [43] |
Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, Fu Q, Mittnik A, Bánffy E, Economou C, Francken M, Friederich S, Pena RG, Hallgren F, Khartanovich V, Khokhlov A, Kunst M, Kuznetsov P, Meller H, Mochalov O, Moiseyev V, Nicklisch N, Pichler SL, Risch R, Rojo Guerra MA, Roth C, Szécsényi-Nagy A, Wahl J, Meyer M, Krause J, Brown D, Anthony D, Cooper A, Alt KW, Reich D (2015) Massive migration from the steppe was a source for Indo-European languages in Europe. Nature, 522, 207- 211.
DOI URL |
| [44] | Hänni C, Laudet V, Sakka M, Bègue A, Stéhelin D (1990) Amplification of mitochondrial DNA fragments from ancient human teeth and bones. Clinical Rheumatology, 310, 365-370. |
| [45] | Hecker HM (1982) Domestication revisited: Its implications for faunal analysis. Journal of Field Archaeology, 9, 217-236. |
| [46] |
Ho SYW, Duchêne S (2014) Molecular-clock methods for estimating evolutionary rates and timescales. Molecular Ecology, 23, 5947-5965.
DOI URL |
| [47] |
Jing Y, Flad RK (2002) Pig domestication in ancient China. Antiquity, 76, 724-732.
DOI URL |
| [48] |
Jones RK, Piper PJ, Wood R, Nguyen AT, Oxenham MF (2019) The Neolithic transition in Vietnam: Assessing evidence for early pig management and domesticated dog. Journal of Archaeological Science: Reports, 28, 102042.
DOI URL |
| [49] |
Kang L, Zheng HX, Zhang M, Yan S, Li L, Liu L, Liu K, Hu K, Chen F, Ma L, Qin Z, Wang Y, Wang X, Jin L (2016) mtDNA analysis reveals enriched pathogenic mutations in Tibetan highlanders. Scientific Reports, 6, 31083.
DOI URL |
| [50] | Katzenberg MA (2008) Stable isotope analysis: A tool for studying past diet, demography, and life history. In: Biological Anthropology of the Human Skeleton (eds Katzenberg MA, Saunders SR), pp. 411-441. John Wiley & Sons, Hoboken, New Jersey. |
| [51] |
Laland KN, Odling-Smee J, Myles S (2010) How culture shaped the human genome: Bringing genetics and the human sciences together. Nature Reviews Genetics, 11, 137-148.
DOI PMID |
| [52] |
Larson G, Burger J (2013) A population genetics view of animal domestication. Trends in Genetics, 29, 197-205.
DOI URL |
| [53] | Larson G, Cucchi T, Fujita M, Matisoo-Smith E, Robins J, Anderson A, Rolett B, Spriggs M, Dolman G, Kim TH, Thuy NTD, Randi E, Doherty M, Due RA, Bollt R, Djubiantono T, Griffin B, Intoh M, Keane E, Kirch P, Li KT, Morwood M, Pedriña LM, Piper PJ, Rabett RJ, Shooter P, van den Bergh G, West E, Wickler S, Yuan J, Cooper A, Dobney K (2007) Phylogeny and ancient DNA of Sus provides insights into Neolithic expansion in Island Southeast Asia and Oceania. Proceedings of the National Academy of Sciences, USA, 104, 4834-4839. |
| [54] |
Larson G, Dobney K, Albarella U, Fang MY, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E, Rowley-Conwy P, Andersson L, Cooper A (2005) Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science, 307, 1618-1621.
DOI URL |
| [55] |
Larson G, Fuller DQ (2014) The evolution of animal domestication. Annual Review of Ecology, Evolution, and Systematics, 45, 115-136.
DOI URL |
| [56] | Larson G, Liu RR, Zhao XB, Yuan J, Fuller D, Barton L, Dobney K, Fan QP, Gu ZL, Liu XH, Luo YB, Lv P, Andersson L, Li N (2010) Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proceedings of the National Academy of Sciences, USA, 107, 7686-7691. |
| [57] | Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M, Barton L, Vigueira CC, Denham T, Dobney K, Doust AN, Gepts P, Gilbert MTP, Gremillion KJ, Lucas L, Lukens L, Marshall FB, Olsen KM, Pires JC, Richerson PJ, de Casas RR, Sanjur OI, Thomas MG, Fuller DQ (2014) Current perspectives and the future of domestication studies. Proceedings of the National Academy of Sciences, USA, 111, 6139-6146. |
| [58] |
Lawler A (2012) In search of the wild chicken. Science, 338, 1020-1024.
DOI URL |
| [59] |
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM (2008) Worldwide human relationships inferred from genome-wide patterns of variation. Science, 319, 1100-1104.
DOI URL |
| [60] | Li LP, Chen NB, Lei CZ (2020) Advance in research on whole-genome genetic diversity and origins in cattle. Animal Husbandry & Veterinary Medicine, 52, 134-138. (in Chinese with English abstract) |
| [ 李联萍, 陈宁博, 雷初朝 (2020) 家牛全基因组遗传多样性与起源研究进展. 畜牧与兽医, 52, 134-138.] | |
| [61] |
Liu L, Bosse M, Megens HJ, Frantz LAF, Lee YL, Irving-Pease EK, Narayan G, Groenen MAM, Madsen O (2019) Genomic analysis on pygmy hog reveals extensive interbreeding during wild boar expansion. Nature Communications, 10, 1992.
DOI URL |
| [62] |
Liu YP, Wu GS, Yao YG, Miao YW, Luikart G, Baig M, Beja- Pereira A, Ding ZL, Palanichamy MG, Zhang YP (2006) Multiple maternal origins of chickens: Out of the Asian jungles. Molecular Phylogenetics and Evolution, 38, 12-19.
DOI URL |
| [63] | Loftus RT, MacHugh DE, Bradley DG, Sharp PM, Cunningham P (1994) Evidence for two independent domestications of cattle. Proceedings of the National Academy of Sciences, USA, 91, 2757-2761. |
| [64] |
Losey RJ (2021) Domestication is not an ancient moment of selection for prosociality: Insights from dogs and modern humans. Journal of Social Archaeology, doi: 10.1177/14696053211055475.
DOI |
| [65] | Luo YB (2012) The Domestication, Raising and Ritual Use of Pigs in Ancient China. Science Press, Beijing. (in Chinese) |
| [ 罗运兵 (2012) 中国古代猪类驯化、饲养与仪式性使用. 科学出版社, 北京.] | |
| [66] | Luo YB (2015) Early development of domestic pig breeding in China. China Cultural Heritage, (4), 40-45. (in Chinese) |
| [ 罗运兵 (2015) 中国家猪饲养的早期发展. 中国文化遗产, (4), 40-45.] | |
| [67] | Luo YB, Li XS (2012) Research on the origin of Chinese domestic pig. Ancient and Modern Agriculture, (3), 10-17. (in Chinese with English abstract) |
| [ 罗运兵, 李想生 (2012) 中国家猪起源机制蠡测. 古今农业, (3), 10-17.] | |
| [68] | Luo YB, Zhang JZ (2008) Restudy of the pigs’ bones from the Jiahu site in Wuyang County, Henan. Archaeology, (1), 90-96. (in Chinese with English abstract) |
| [ 罗运兵, 张居中 (2008) 河南舞阳县贾湖遗址出土猪骨的再研究. 考古, (1), 90-96.] | |
| [69] |
Lv FH, Peng WF, Yang J, Zhao YX, Li WR, Liu MJ, Ma YH, Zhao QJ, Yang GL, Wang F, Li JQ, Liu YG, Shen ZQ, Zhao SG, Hehua EE, Gorkhali NA, Farhad Vahidi SM, Muladno M, Naqvi AN, Tabell J, Iso-Touru T, Bruford MW, Kantanen J, Han JL, Li MH (2015) Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Molecular Biology and Evolution, 32, 2515-2533.
DOI URL |
| [70] |
MacHugh DE, Larson G, Orlando L (2017) Taming the past: Ancient DNA and the study of animal domestication. Annual Review of Animal Biosciences, 5, 329-351.
DOI PMID |
| [71] |
Maricic T, Whitten M, Pääbo S (2010) Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE, 5, e14004.
DOI URL |
| [72] | Mason IL (1984) Evolution of Domesticated Animals. Longman Group, London. |
| [73] |
Miao YW, Peng MS, Wu GS, Ouyang YN, Yang ZY, Yu N, Liang JP, Pianchou G, Beja-Pereira A, Mitra B, Palanichamy MG, Baig M, Chaudhuri TK, Shen YY, Kong QP, Murphy RW, Yao YG, Zhang YP (2013) Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity, 110, 277-282.
DOI PMID |
| [74] |
Mourier T, Ho SYW, Gilbert MTP, Willerslev E, Orlando L (2012) Statistical guidelines for detecting past population shifts using ancient DNA. Molecular Biology and Evolution, 29, 2241-2251.
DOI PMID |
| [75] | Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, USA, 76, 5269-5273. |
| [76] |
Orellana-González E, Sparacello VS, Bocaege E, Varalli A, Moggi-Cecchi J, Dori I (2020) Insights on patterns of developmental disturbances from the analysis of linear enamel hypoplasia in a Neolithic sample from Liguria (northwestern Italy). International Journal of Paleopathology, 28, 123-136.
DOI PMID |
| [77] | Orlando L (2020a) Ancient genomes reveal unexpected horse domestication and management dynamics. BioEssays, 42, e1900164. |
| [78] |
Orlando L (2020b) The evolutionary and historical foundation of the modern horse: Lessons from ancient genomics. Annual Review of Genetics, 54, 563-581.
DOI URL |
| [79] |
Orlando L, Cooper A (2014) Using ancient DNA to understand evolutionary and ecological processes. Annual Review of Ecology, Evolution, and Systematics, 45, 573-598.
DOI URL |
| [80] |
Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PLF, Fumagalli M, Vilstrup JT, Raghavan M, Korneliussen T, Malaspinas AS, Vogt J, Szklarczyk D, Kelstrup CD, Vinther J, Dolocan A, Stenderup J, Velazquez AMV, Cahill J, Rasmussen M, Wang X, Min J, Zazula GD, Seguin-Orlando A, Mortensen C, Magnussen K, Thompson JF, Weinstock J, Gregersen K, Røed KH, Eisenmann V, Rubin CJ, Miller DC, Antczak DF, Bertelsen MF, Brunak S, Al-Rasheid KAS, Ryder O, Andersson L, Mundy J, Krogh A, Gilbert MTP, Kjær K, Sicheritz-Ponten T, Jensen LJ, Olsen JV, Hofreiter M, Nielsen R, Shapiro B, Wang J, Willerslev E (2013) Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature, 499, 74-78.
DOI URL |
| [81] | Ormond L, Foll M, Ewing GB, Pfeifer SP, Jensen JD (2016) Inferring the age of a fixed beneficial allele. Molecular Ecology, 25, 157-169. |
| [82] |
Pääbo S, Higuchi RG, Wilson AC (1989) Ancient DNA and the polymerase chain reaction. The emerging field of molecular archaeology. Journal of Biological Chemistry, 264, 9709-9712.
PMID |
| [83] |
Park SDE, Magee DA, McGettigan PA, Teasdale MD, Edwards CJ, Lohan AJ, Murphy A, Braud M, Donoghue MT, Liu Y, Chamberlain AT, Rue-Albrecht K, Schroeder S, Spillane C, Tai SS, Bradley DG, Sonstegard TS, Loftus BJ, MacHugh DE (2015) Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biology, 16, 234.
DOI URL |
| [84] |
Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan YP, Genschoreck T, Webster T, Reich D (2012) Ancient admixture in human history. Genetics, 192, 1065-1093.
DOI PMID |
| [85] | Peters J, Lebrasseur O, Best J, Miller H, Fothergill T, Dobney K, Thomas RM, Maltby M, Sykes N, Hanotte O, O’Connor T, Collins MJ, Larson G (2015) Questioning new answers regarding Holocene chicken domestication in China. Proceedings of the National Academy of Sciences, USA, 112, E2415. |
| [86] |
Pinhasi R, Fernandes D, Sirak K, Novak M, Connell S, Alpaslan-Roodenberg S, Gerritsen F, Moiseyev V, Gromov A, Raczky P, Anders A, Pietrusewsky M, Rollefson G, Jovanovic M, Trinhhoang H, Bar-Oz G, Oxenham M, Matsumura H, Hofreiter M (2015) Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS ONE, 10, e0129102.
DOI URL |
| [87] |
Pitt D, Sevane N, Nicolazzi EL, MacHugh DE, Park SDE, Colli L, Martinez R, Bruford MW, Orozco-Terwengel P (2019) Domestication of cattle: Two or three events? Evolutionary Applications, 12, 123-136.
DOI URL |
| [88] |
Pitulko VV, Kasparov AK (2017) Archaeological dogs from the Early Holocene Zhokhov site in the Eastern Siberian Arctic. Journal of Archaeological Science: Reports, 13, 491- 515.
DOI URL |
| [89] | Porter V (1993) Pigs: A Handbook to the Breeds of the World. Comstock Publishing Associates. Cornell University Press, Ithaca, New York. |
| [90] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
DOI PMID |
| [91] |
Reich D, Thangaraj K, Patterson N, Price AL, Singh L (2009) Reconstructing Indian population history. Nature, 461, 489-494.
DOI URL |
| [92] |
Rowley-Conwy P, Albarella U, Dobney K (2012) Distinguishing wild boar from domestic pigs in prehistory: A review of approaches and recent results. Journal of World Prehistory, 25, 1-44.
DOI URL |
| [93] |
Sabatini S, Bergerbrant S, Brandt LØ, Margaryan A, Allentoft ME (2019) Approaching sheep herds origins and the emergence of the wool economy in continental Europe during the Bronze Age. Archaeological and Anthropological Sciences, 11, 4909-4925.
DOI |
| [94] | Sánchez-Quinto F, Lalueza-Fox C (2015) Almost 20 years of Neanderthal palaeogenetics: Adaptation, admixture, diversity, demography and extinction. Philosophical Transactions of the Royal Society B: Biological Sciences, 370, 20130374. |
| [95] |
Soraggi S, Wiuf C, Albrechtsen A (2018) Powerful inference with the D-statistic on low-coverage whole-genome data. G3 Genes|Genomes|Genetics, 8, 551-566.
DOI URL |
| [96] |
Sunnåker M, Busetto AG, Numminen E, Corander J, Foll M, Dessimoz C (2013) Approximate Bayesian computation. PLoS Computational Biology, 9, e1002803.
DOI URL |
| [97] |
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585-595.
DOI PMID |
| [98] | Tao KT (2021) Study on Genetic Diversity of Asian Domestic Horse--Examples from Chinese and Mongolian Populations. PhD dissertation, Inner Mongolia Agricultural University, Hohhot. (in Chinese with English abstract) |
| [ 陶克涛 (2021) 亚洲家马遗传多样性的研究--以中国和蒙古国种群为例. 博士学位论文, 内蒙古农业大学, 呼和浩特.] | |
| [99] | Teletchea F (2019) A brief overview. In: Animal Domestication (ed. Teletchea F). IntechOpen, London. |
| [100] |
Terrell JE, Hart JP, Barut S, Cellinese N, Curet A, Denham T, Kusimba CM, Latinis K, Oka R, Palka J, Pohl MED, Pope KO, Williams PR, Haines H, Staller JE (2003) Domesticated landscapes: The subsistence ecology of plant and animal domestication. Journal of Archaeological Method and Theory, 10, 323-368.
DOI URL |
| [101] |
Tong X, Hou L, He W, Mei C, Huang B, Zhang C, Hu C, Wang C (2020) Whole genome sequence analysis reveals genetic structure and X-chromosome haplotype structure in indigenous Chinese pigs. Scientific Reports, 10, 9433.
DOI URL |
| [102] |
Varela MA, Amos W (2010) Heterogeneous distribution of SNPs in the human genome: Microsatellites as predictors of nucleotide diversity and divergence. Genomics, 95, 151- 159.
DOI URL |
| [103] |
Wang C, Wang HY, Zhang Y, Tang ZL, Li K, Liu B (2015) Genome-wide analysis reveals artificial selection on coat colour and reproductive traits in Chinese domestic pigs. Molecular Ecology Resources, 15, 414-424.
DOI URL |
| [104] |
Wang GD, Xie HB, Peng MS, Irwin D, Zhang YP (2014) Domestication genomics: Evidence from animals. Annual Review of Animal Biosciences, 2, 65-84.
DOI URL |
| [105] |
Wang GD, Zhai W, Yang HC, Wang L, Zhong L, Liu YH, Fan RX, Yin TT, Zhu CL, Poyarkov AD, Irwin DM, Hytönen MK, Lohi H, Wu CI, Savolainen P, Zhang YP (2016) Out of southern East Asia: The natural history of domestic dogs across the world. Cell Research, 26, 21-33.
DOI URL |
| [106] |
Wang H, Sofer T, Zhang X, Elston RC, Redline S, Zhu X (2020a) Local ancestry inference in large pedigrees. Scientific Reports, 10, 189.
DOI URL |
| [107] |
Wang MS, Thakur M, Peng MS, Jiang Y, Frantz LAF, Li M, Zhang JJ, Wang S, Peters J, Otecko NO, Suwannapoom C, Guo X, Zheng ZQ, Esmailizadeh A, Hirimuthugoda NY, Ashari H, Suladari S, Zein MSA, Kusza S, Sohrabi S, Kharrati-Koopaee H, Shen QK, Zeng L, Yang MM, Wu YJ, Yang XY, Lu XM, Jia XZ, Nie QH, Lamont SJ, Lasagna E, Ceccobelli S, Gunwardana HGTN, Senasige TM, Feng SH, Si JF, Zhang H, Jin JQ, Li ML, Liu YH, Chen HM, Ma C, Dai SS, Bhuiyan AKFH, Khan MS, Silva GLLP, Le TT, Mwai OA, Ibrahim MNM, Supple M, Shapiro B, Hanotte O, Zhang GJ, Larson G, Han JL, Wu DD, Zhang YP (2020b) 863 genomes reveal the origin and domestication of chicken. Cell Research, 30, 693-701.
DOI URL |
| [108] |
Wang MS, Zhang JJ, Guo X, Li M, Meyer R, Ashari H, Zheng ZQ, Wang S, Peng MS, Jiang Y, Thakur M, Suwannapoom C, Esmailizadeh A, Hirimuthugoda NY, Zein MSA, Kusza S, Kharrati-Koopaee H, Zeng L, Wang YM, Yin TT, Yang MM, Li ML, Lu XM, Lasagna E, Ceccobelli S, Gunwardana HGTN, Senasig TM, Feng SH, Zhang H, Bhuiyan AKFH, Khan MS, Silva GLLP, Thuy LT, Mwai OA, Ibrahim MNM, Zhang GJ, Qu KX, Hanotte O, Shapiro B, Bosse M, Wu DD, Han JL, Zhang YP (2021) Large-scale genomic analysis reveals the genetic cost of chicken domestication. BMC Biology, 19, 118.
DOI URL |
| [109] | Wen ZL, Zhao YQ (2021) Progress on animal domestication under population genetics. Hereditas (Beijing), 43, 226-239. (in Chinese with English abstract) |
| [ 文子龙, 赵毅强 (2021) 群体遗传学下动物驯化研究进展. 遗传, 43, 226-239.] | |
| [110] |
West B, Zhou BX (1988) Did chickens go North? New evidence for domestication. Journal of Archaeological Science, 15, 515-533.
DOI URL |
| [111] |
White S (2011) From globalized pig breeds to capitalist pigs: A study in animal cultures and evolutionary history. Environmental History, 16, 94-120.
DOI URL |
| [112] |
Willerslev E, Hansen AJ, Binladen J, Brand TB, Gilbert MTP, Shapiro B, Bunce M, Wiuf C, Gilichinsky DA, Cooper A (2003) Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science, 300, 791-795.
PMID |
| [113] | Wing E (2011) Animal domestication in the Andes. In: Advances in Andean Archaeology (ed. David LB), pp. 167-188. De Gruyter Mouton, Berlin. |
| [114] |
Wu GS, Yao YG, Qu KX, Ding ZL, Li H, Palanichamy MG, Duan ZY, Li N, Chen YS, Zhang YP (2007) Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biology, 8, R245.
DOI URL |
| [115] |
Wu J, Liu YX, Zhao YQ (2021) Systematic review on local ancestor inference from a mathematical and algorithmic perspective. Frontiers in Genetics, 12, 639877.
DOI URL |
| [116] | Xiang H, Gao JQ, Yu BQ, Zhou H, Cai DW, Zhang YW, Chen XY, Wang X, Hofreiter M, Zhao XB (2014) Early Holocene chicken domestication in Northern China. Proceedings of the National Academy of Sciences, USA, 111, 17564-17569. |
| [117] | Yuan J (2015) Zooarchaeology of China. Cultural Relics Publishing House, Beijing. (in Chinese with English abstract) |
| [ 袁靖 (2015) 中国动物考古学. 文物出版社, 北京.] | |
| [118] | Yuan J, Dong NN (2018) The reexamination on the origination of the domestication of animals in China. Archaeology, (9), 113-120, 2. (in Chinese with English abstract) |
| [ 袁靖, 董宁宁 (2018) 中国家养动物起源的再思考. 考古, (9), 113-120, 2.] | |
| [119] | Yuan J, Lu P, Li ZP, Deng H, Jiangtian M (2015) Restudy of the origin of domestic chicken in ancient China. Cultural Relics in Southern China, (3), 53-57. (in Chinese with English abstract) |
| [ 袁靖, 吕鹏, 李志鹏, 邓惠, 江田真毅 (2015) 中国古代家鸡起源的再研究. 南方文物, (3), 53-57.] | |
| [120] |
Yurtman E, Özer O, Yüncü E, Dağtaş ND, Koptekin D, Çakan YG, Özkan M, Akbaba A, Kaptan D, Atağ G, Vural KB, Gündem CY, Martin L, Kılınç GM, Ghalichi A, Açan SC, Yaka R, Sağlıcan E, Lagerholm VK, Krzewińska M, Günther T, Morell Miranda P, Pişkin E, Şevketoğlu M, Bilgin CC, Atakuman Ç, Erdal YS, Sürer E, Altınışık NE, Lenstra JA, Yorulmaz S, Abazari MF, Hoseinzadeh J, Baird D, Bıçakçı E, Çevik Ö, Gerritsen F, Özbal R, Götherström A, Somel M, Togan İ, Özer F (2021) Archaeogenetic analysis of Neolithic sheep from Anatolia suggests a complex demographic history since domestication. Communications Biology, 4, 1279.
DOI PMID |
| [121] | Zeder MA (2008) Domestication and early agriculture in the Mediterranean Basin:Origins, diffusion, and impact. Proceedings of the National Academy of Sciences, USA, 105, 11597-11604. |
| [122] |
Zeder MA (2012) The domestication of animals. Journal of Anthropological Research, 68, 161-190.
DOI URL |
| [123] | Zeder MA (2015) Core questions in domestication research. Proceedings of the National Academy of Sciences, USA, 112, 3191-3198. |
| [124] |
Zeder MA, Smith BD (2009) A conversation on agricultural origins. Current Anthropology, 50, 681-690.
DOI URL |
| [125] | Zelditch ML, Swiderski DL, Sheets HD, Fink WL (2004) Geometric Morphometrics for Biologists. Academic Press, San Diego. |
| [126] |
Zeuner FE (1964) A history of domesticated animals. Journal of Animal Ecology, 33, 215.
DOI URL |
| [127] |
Zhang Z, Jia Y, Chen Y, Wang L, Lv X, Yang F, He Y, Ning Z, Qu L (2018) Genomic variation in Pekin duck populations developed in three different countries as revealed by whole-genome data. Animal Genetics, 49, 132-136.
DOI PMID |
| [128] |
Zhao YX, Yang J, Lv FH, Hu XJ, Xie XL, Zhang M, Li WR, Liu MJ, Wang YT, Li JQ, Liu YG, Ren YL, Wang F, Hehua EE, Kantanen J, Arjen Lenstra J, Han JL, Li MH (2017) Genomic reconstruction of the history of native sheep reveals the peopling patterns of nomads and the expansion of early pastoralism in East Asia. Molecular Biology and Evolution, 34, 2380-2395.
DOI URL |
| [129] | Zheng ZQ (2019) Population structure and genetic introgression from wild relatives in worldwide goat populations. PhD dissertation. Northwest A&F University, Yangling. (in Chinese with English abstract) |
| [ 郑竹清 (2019) 世界山羊群体遗传结构及其野生近缘种基因渗入研究. 博士学位论文, 西北农林科技大学, 杨凌.] | |
| [130] | Zhu WB, Chen YF (2018) Meat consumption and outlook in the world and China. Agricultural Outlook, 14, 98-109. (in Chinese with English abstract) |
| [ 朱文博, 陈永福 (2018) 世界和中国肉类消费及展望. 农业展望, 14, 98-109.] |
| [1] | 陶克涛, 韩海格, 赵若阳, 图格琴, 芒来, 白东义. 家马的驯化起源与遗传演化特征[J]. 生物多样性, 2020, 28(6): 734-748. |
| [2] | 张文驹, 戎俊, 韦朝领, 高连明, 陈家宽. 栽培茶树的驯化起源与传播[J]. 生物多样性, 2018, 26(4): 357-372. |
| [3] | 孙立夫, 裴克全, 张艳华, 赵俊, 杨国亭, 秦国夫, 宋玉双, 宋瑞清. 中国与欧洲高卢蜜环菌的遗传多样性[J]. 生物多样性, 2012, 20(2): 224-230. |
| [4] | 徐福荣, 张恩来, 董超, 戴陆园, 张红生. 云南元阳哈尼梯田两个不同时期种植的水稻地方品种表型比较[J]. 生物多样性, 2010, 18(4): 365-372. |
| [5] | 张丽娜, 叶武威, 王俊娟, 樊保香, 王德龙. 棉花耐盐相关种质资源遗传多样性分析[J]. 生物多样性, 2010, 18(2): 137-144. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn