



生物多样性 ›› 2008, Vol. 16 ›› Issue (1): 53-62. DOI: 10.3724/SP.J.1003.2008.07050 cstr: 32101.14.SP.J.1003.2008.07050
王建民1,2, 岳文斌1,*
收稿日期:2007-02-15
接受日期:2007-07-30
出版日期:2008-01-27
发布日期:2008-01-27
通讯作者:
岳文斌
作者简介:*E-mail:wb-yue@126.com基金资助:Jianmin Wang1,2, Wenbin Yue1,*
Received:2007-02-15
Accepted:2007-07-30
Online:2008-01-27
Published:2008-01-27
Contact:
Wenbin Yue
摘要:
为了探讨家养绵羊与山羊的属间遗传关系, 我们利用13个定位于绵羊染色体上的微卫星基因座, 分析了黄河下游4个地方绵羊品种、4个地方山羊品种和1个杂交绵羊类群的遗传结构及其系统发生关系。经Hardy-Weinberg平衡检验和中性测试, 发现地方绵羊和山羊种群均处于不平衡状态(P< 0.01), 61.53%的基因座属于中性位点, 说明所研究种群属于非随机交配, 可能受到选择、迁移等进化因素的影响。对有效等位基因数、多态信息含量、Shannon信息指数、观察杂合度和期望杂合度等遗传多样性参数进行比较, 发现绵羊种群的遗传变异程度明显(P<0.01或P<0.05)高于山羊种群, 但不同基因座上的差异效应不一致; 结合F统计量和亲缘关系等参数, 可推测绵羊和山羊虽然均存在不同程度的近交现象(He>Ho, FIS>0),但分别属于杂交和近交繁殖。通过群体遗传分化和系统发育拓扑结构分析, 证明绵羊属由共同祖先分化而来的时间晚于山羊属, 两属间的遗传距离为1.0708-1.5927, 遗传分化时间约为19,807-28,955年; 绵羊属内品种间的遗传分化程度(FST<0.05)均低于山羊属内品种间的分化(FST>0.15)。本研究揭示了人工选择对同域家养绵羊与山羊交配系统的形成及群体遗传分化具有深刻的影响。
王建民, 岳文斌 (2008) 黄河下游家绵羊与家山羊遗传关系的微卫星分析. 生物多样性, 16, 53-62. DOI: 10.3724/SP.J.1003.2008.07050.
Jianmin Wang, Wenbin Yue (2008) Genetic relationships of domestic sheep and goats in the lower reaches of the Yellow River based on microsatellite analysis. Biodiversity Science, 16, 53-62. DOI: 10.3724/SP.J.1003.2008.07050.
| 品种名称 Breeds | 采样地点 Sampling location | 采样只数(羊群数) Sample size (flock number) |
|---|---|---|
| 绵羊品种 Native or crossbred sheep breeds | ||
| 小尾寒羊 Small-tailed Han (STH) | 山东嘉祥县 Jiaxiang County | 42 (14) |
| 大尾寒羊 Large-tailed Han (LTH) | 山东临清市 Linqing City | 42 (22) |
| 山地绵羊 Shandi (SD) | 山东莱芜市 Laiwu City | 29 (17) |
| 洼地绵羊 Wadi (WD) | 山东滨州市 Binzhou City | 40 (18) |
| 杂交绵羊 Crossbred (CB) | 山东莱芜市 Laiwu City | 20 (1) |
| 山羊品种 Native goat breeds | ||
| 济宁青山羊 Jining grey (JNG) | 山东嘉祥县 Jiaxiang County | 40 (5) |
| 鲁北白山羊 Lubei white (LBW) | 山东无棣县 Wudi County | 40 (23) |
| 莱芜黑山羊 Laiwu black (LWB) | 山东莱芜市 Laiwu City | 40 (26) |
| 崂山奶山羊 Laoshan milk (LSM) | 山东临朐县 Linqu County | 40 (21) |
表1 所研究山东省羊品种的名称、采样地点和样本数量
Table 1 Names, sampling localities and sample sizes of studied breeds from Shandong Province
| 品种名称 Breeds | 采样地点 Sampling location | 采样只数(羊群数) Sample size (flock number) |
|---|---|---|
| 绵羊品种 Native or crossbred sheep breeds | ||
| 小尾寒羊 Small-tailed Han (STH) | 山东嘉祥县 Jiaxiang County | 42 (14) |
| 大尾寒羊 Large-tailed Han (LTH) | 山东临清市 Linqing City | 42 (22) |
| 山地绵羊 Shandi (SD) | 山东莱芜市 Laiwu City | 29 (17) |
| 洼地绵羊 Wadi (WD) | 山东滨州市 Binzhou City | 40 (18) |
| 杂交绵羊 Crossbred (CB) | 山东莱芜市 Laiwu City | 20 (1) |
| 山羊品种 Native goat breeds | ||
| 济宁青山羊 Jining grey (JNG) | 山东嘉祥县 Jiaxiang County | 40 (5) |
| 鲁北白山羊 Lubei white (LBW) | 山东无棣县 Wudi County | 40 (23) |
| 莱芜黑山羊 Laiwu black (LWB) | 山东莱芜市 Laiwu City | 40 (26) |
| 崂山奶山羊 Laoshan milk (LSM) | 山东临朐县 Linqu County | 40 (21) |
| 基因座 Loci | Na | Ne | Dk | PIC | I | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | |||||
| MAF70 | 12.00a | 6.75b | 6.10 | 4.52 | 0.83 | 0.79 | 0.81 | 0.74 | 2.06a | 1.60b | ||||
| BMS1248 | 9.80 | 6.25 | 5.28 | 3.91 | 0.83 | 0.74 | 0.75 | 0.69 | 1.85 | 1.52 | ||||
| BM6404 | 11.60 | 6.50 | 5.28 | 3.81 | 0.84 | 0.73 | 0.77 | 0.68 | 1.88 | 1.49 | ||||
| BMS1714 | 10.40a | 4.67b | 5.97 | 2.94 | 0.83 | 0.72 | 0.79 | 0.65 | 1.94 | 1.35 | ||||
| BMS1724 | 12.60A | 2.67B | 8.55A | 1.98B | 0.91A | 0.62B | 0.80a | 0.52b | 2.27A | 0.96B | ||||
| BMS875 | 9.00a | 3.00b | 4.54a | 1.98b | 0.80a | 0.60b | 0.74a | 0.50b | 1.72A | 0.96B | ||||
| BMS1678 | 10.80a | 6.00b | 7.29a | 3.51b | 0.89a | 0.69b | 0.83 | 0.62 | 2.10a | 1.37b | ||||
| BM203 | 13.40A | 1.75B | 8.44A | 1.62B | 0.91A | 0.46B | 0.85A | 0.26B | 2.26A | 0.48B | ||||
| MB023 | 11.80A | 2.67B | 7.66A | 1.70B | 0.88A | 0.56B | 0.85A | 0.45B | 2.20A | 0.84B | ||||
| BM3033 | 12.80A | 6.25B | 9.41A | 4.79B | 0.92A | 0.80B | 0.88A | 0.76B | 2.33A | 1.65B | ||||
| BMC1206 | 8.20A | 3.50B | 5.56A | 2.44B | 0.83a | 0.57b | 0.79A | 0.50B | 1.84A | 0.98B | ||||
| BM6526 | 11.60a | 4.25b | 6.79 | 2.86 | 0.88a | 0.60b | 0.82a | 0.54b | 2.09A | 1.11B | ||||
| BM6444 | 4.20b | 7.00a | 2.51 | 4.60 | 0.61 | 0.70 | 0.54 | 0.66 | 1.07 | 1.54 | ||||
| 平均 Average | 10.63A | 4.77B | 6.41A | 3.36B | 0.84A | 0.66B | 0.79A | 0.58B | 1.97A | 1.23B | ||||
表2 绵羊和山羊种群在13个基因座上的等位基因数(Na)、有效等位基因数(Ne)、基因多样度(Dk)、多态信息含量(PIC)和Shannon指数(I)
Table 2 Number of alleles (Na), number of effective alleles (Ne), gene diversity (Dk), polymorphism information content (PIC) and Shannon's information index (I) of 13 loci in sheep and goat populations
| 基因座 Loci | Na | Ne | Dk | PIC | I | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | |||||
| MAF70 | 12.00a | 6.75b | 6.10 | 4.52 | 0.83 | 0.79 | 0.81 | 0.74 | 2.06a | 1.60b | ||||
| BMS1248 | 9.80 | 6.25 | 5.28 | 3.91 | 0.83 | 0.74 | 0.75 | 0.69 | 1.85 | 1.52 | ||||
| BM6404 | 11.60 | 6.50 | 5.28 | 3.81 | 0.84 | 0.73 | 0.77 | 0.68 | 1.88 | 1.49 | ||||
| BMS1714 | 10.40a | 4.67b | 5.97 | 2.94 | 0.83 | 0.72 | 0.79 | 0.65 | 1.94 | 1.35 | ||||
| BMS1724 | 12.60A | 2.67B | 8.55A | 1.98B | 0.91A | 0.62B | 0.80a | 0.52b | 2.27A | 0.96B | ||||
| BMS875 | 9.00a | 3.00b | 4.54a | 1.98b | 0.80a | 0.60b | 0.74a | 0.50b | 1.72A | 0.96B | ||||
| BMS1678 | 10.80a | 6.00b | 7.29a | 3.51b | 0.89a | 0.69b | 0.83 | 0.62 | 2.10a | 1.37b | ||||
| BM203 | 13.40A | 1.75B | 8.44A | 1.62B | 0.91A | 0.46B | 0.85A | 0.26B | 2.26A | 0.48B | ||||
| MB023 | 11.80A | 2.67B | 7.66A | 1.70B | 0.88A | 0.56B | 0.85A | 0.45B | 2.20A | 0.84B | ||||
| BM3033 | 12.80A | 6.25B | 9.41A | 4.79B | 0.92A | 0.80B | 0.88A | 0.76B | 2.33A | 1.65B | ||||
| BMC1206 | 8.20A | 3.50B | 5.56A | 2.44B | 0.83a | 0.57b | 0.79A | 0.50B | 1.84A | 0.98B | ||||
| BM6526 | 11.60a | 4.25b | 6.79 | 2.86 | 0.88a | 0.60b | 0.82a | 0.54b | 2.09A | 1.11B | ||||
| BM6444 | 4.20b | 7.00a | 2.51 | 4.60 | 0.61 | 0.70 | 0.54 | 0.66 | 1.07 | 1.54 | ||||
| 平均 Average | 10.63A | 4.77B | 6.41A | 3.36B | 0.84A | 0.66B | 0.79A | 0.58B | 1.97A | 1.23B | ||||
| 基因座 Loci | Ho | He | FST | FIS | Relat | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | |||||
| MAF70 | 0.599A | 0.225B | 0.850 | 0.783 | 0.017 | 0.084 | 0.330 | 0.715 | 0.025 | 0.096 | ||||
| BMS1248 | 0.401 | 0.400 | 0.796 | 0.737 | 0.061 | 0.066 | 0.543 | 0.460 | 0.078 | 0.088 | ||||
| BM6404 | 0.489B | 0.850A | 0.811 | 0.727 | 0.006 | 0.158 | 0.487 | -0.172 | 0.008 | 0.311 | ||||
| BMS1714 | 0.395 | 0.731 | 0.839 | 0.538 | 0.046 | 0.091 | 0.622 | -0.364 | 0.057 | 0.239 | ||||
| BMS1724 | 0.419 | 0.281 | 0.896a | 0.460b | 0.024 | 0.392 | 0.538 | 0.121 | 0.032 | -1.288 | ||||
| BMS875 | 0.361 | 0.256 | 0.787a | 0.447b | 0.052 | 0.070 | 0.474 | 0.430 | 0.069 | 0.096 | ||||
| BMS1678 | 0.525 | 0.563 | 0.875A | 0.689B | 0.022 | 0.204 | 0.377 | 0.185 | 0.031 | 0.301 | ||||
| BM203 | 0.631A | 0.219B | 0.884A | 0.340B | 0.013 | 0.359 | 0.375 | 0.360 | 0.019 | 0.451 | ||||
| MB023 | 0.699A | 0.031B | 0.889A | 0.414B | 0.034 | 0.371 | 0.256 | 0.925 | 0.054 | 0.379 | ||||
| BM3033 | 0.804 | 0.856 | 0.913A | 0.799B | 0.002 | 0.065 | 0.132 | -0.073 | 0.003 | 0.130 | ||||
| BMC1206 | 0.583A | 0.000B | 0.837A | 0.566B | 0.021 | 0.122 | 0.338 | 1.000 | 0.031 | 0.122 | ||||
| BM6526 | 0.540 | 0.488 | 0.869a | 0.595b | 0.012 | 0.294 | 0.370 | 0.183 | 0.017 | 0.413 | ||||
| BM6444 | 0.260B | 0.750A | 0.610 | 0.699 | 0.171 | 0.165 | 0.594 | -0.075 | 0.205 | 0.300 | ||||
| 平均 Average | 0.516 | 0.464 | 0.835A | 0.646B | 0.035 | 0.164 | 0.412 | 0.280 | 0.049b | 0.234a | ||||
表3 绵羊和山羊种群在13个基因座上的观测杂合度(Ho)、期望杂合度(He)、固定指数(FST)、近交系数((FIS)和亲缘关系指数(Relat)
Table 3 Observed and expected heterozygosity (Ho, He), fixation index (FST), inbreeding coefficient (FIS) and relatedness (Relat) of 13 loci in sheep and goat populations
| 基因座 Loci | Ho | He | FST | FIS | Relat | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | 绵羊 Sheep | 山羊 Goat | |||||
| MAF70 | 0.599A | 0.225B | 0.850 | 0.783 | 0.017 | 0.084 | 0.330 | 0.715 | 0.025 | 0.096 | ||||
| BMS1248 | 0.401 | 0.400 | 0.796 | 0.737 | 0.061 | 0.066 | 0.543 | 0.460 | 0.078 | 0.088 | ||||
| BM6404 | 0.489B | 0.850A | 0.811 | 0.727 | 0.006 | 0.158 | 0.487 | -0.172 | 0.008 | 0.311 | ||||
| BMS1714 | 0.395 | 0.731 | 0.839 | 0.538 | 0.046 | 0.091 | 0.622 | -0.364 | 0.057 | 0.239 | ||||
| BMS1724 | 0.419 | 0.281 | 0.896a | 0.460b | 0.024 | 0.392 | 0.538 | 0.121 | 0.032 | -1.288 | ||||
| BMS875 | 0.361 | 0.256 | 0.787a | 0.447b | 0.052 | 0.070 | 0.474 | 0.430 | 0.069 | 0.096 | ||||
| BMS1678 | 0.525 | 0.563 | 0.875A | 0.689B | 0.022 | 0.204 | 0.377 | 0.185 | 0.031 | 0.301 | ||||
| BM203 | 0.631A | 0.219B | 0.884A | 0.340B | 0.013 | 0.359 | 0.375 | 0.360 | 0.019 | 0.451 | ||||
| MB023 | 0.699A | 0.031B | 0.889A | 0.414B | 0.034 | 0.371 | 0.256 | 0.925 | 0.054 | 0.379 | ||||
| BM3033 | 0.804 | 0.856 | 0.913A | 0.799B | 0.002 | 0.065 | 0.132 | -0.073 | 0.003 | 0.130 | ||||
| BMC1206 | 0.583A | 0.000B | 0.837A | 0.566B | 0.021 | 0.122 | 0.338 | 1.000 | 0.031 | 0.122 | ||||
| BM6526 | 0.540 | 0.488 | 0.869a | 0.595b | 0.012 | 0.294 | 0.370 | 0.183 | 0.017 | 0.413 | ||||
| BM6444 | 0.260B | 0.750A | 0.610 | 0.699 | 0.171 | 0.165 | 0.594 | -0.075 | 0.205 | 0.300 | ||||
| 平均 Average | 0.516 | 0.464 | 0.835A | 0.646B | 0.035 | 0.164 | 0.412 | 0.280 | 0.049b | 0.234a | ||||
| 品种 Breeds | 品种 Breeds | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| STH | LTH | WD | SD | CB | JNG | LBW | LSM | LWB | |
| STH | ** | ** | ** | NS | ** | ** | ** | ** | |
| LTH | 0.0196 | ** | ** | NS | ** | ** | ** | ** | |
| WD | 0.0267 | 0.0312 | ** | NS | ** | ** | ** | ** | |
| SD | 0.0405 | 0.0408 | 0.0440 | ** | ** | ** | ** | ** | |
| CB | 0.0212 | 0.0472 | 0.0599 | 0.0595 | ** | ** | ** | ** | |
| JNG | 0.2243 | 0.2126 | 0.2472 | 0.2334 | 0.2791 | ** | ** | ** | |
| LBW | 0.1597 | 0.1549 | 0.1784 | 0.1677 | 0.1790 | 0.1283 | ** | ** | |
| LSM | 0.2131 | 0.2138 | 0.2418 | 0.2292 | 0.2697 | 0.2645 | 0.1579 | ** | |
| LWB | 0.1446 | 0.1400 | 0.1726 | 0.1503 | 0.1612 | 0.1669 | 0.0954 | 0.1535 | |
表4 基于9个基因座的9个品种间成对固定指数(FST, 对角线下方)和遗传分化配对测试(对角线上方)
Table 4 Fixation index (FST, below the diagonal) and pairwise testing of genetic differentiation (above the diagonal) among nine breeds based on nine loci
| 品种 Breeds | 品种 Breeds | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| STH | LTH | WD | SD | CB | JNG | LBW | LSM | LWB | |
| STH | ** | ** | ** | NS | ** | ** | ** | ** | |
| LTH | 0.0196 | ** | ** | NS | ** | ** | ** | ** | |
| WD | 0.0267 | 0.0312 | ** | NS | ** | ** | ** | ** | |
| SD | 0.0405 | 0.0408 | 0.0440 | ** | ** | ** | ** | ** | |
| CB | 0.0212 | 0.0472 | 0.0599 | 0.0595 | ** | ** | ** | ** | |
| JNG | 0.2243 | 0.2126 | 0.2472 | 0.2334 | 0.2791 | ** | ** | ** | |
| LBW | 0.1597 | 0.1549 | 0.1784 | 0.1677 | 0.1790 | 0.1283 | ** | ** | |
| LSM | 0.2131 | 0.2138 | 0.2418 | 0.2292 | 0.2697 | 0.2645 | 0.1579 | ** | |
| LWB | 0.1446 | 0.1400 | 0.1726 | 0.1503 | 0.1612 | 0.1669 | 0.0954 | 0.1535 | |
| 品种 Breeds | 品种 Breeds | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| STH | LTH | WD | SD | CB | JNG | LBW | LSM | LWB | |
| STH | 4,569 | 3,972 | 5,725 | 5,396 | 27,154 | 23,441 | 23,105 | 19,807 | |
| LTH | 0.2513 | 5,229 | 6,174 | 8,405 | 25,063 | 24,943 | 25,639 | 19,467 | |
| WD | 0.2185 | 0.2876 | 5,092 | 8,894 | 28,713 | 22,550 | 24,521 | 24,490 | |
| SD | 0.3149 | 0.3396 | 0.2801 | 9,286 | 28,955 | 24,108 | 25,823 | 21,002 | |
| CB | 0.2968 | 0.4623 | 0.4892 | 0.5108 | 34,493 | 26,606 | 30,001 | 21,022 | |
| JNG | 1.4936 | 1.3786 | 1.5794 | 1.5927 | 1.8973 | 10,626 | 17,940 | 12,901 | |
| LBW | 1.2894 | 1.3720 | 1.2404 | 1.3261 | 1.4635 | 0.5845 | 6,743 | 7,561 | |
| LSM | 1.2709 | 1.4103 | 1.3488 | 1.4204 | 1.6502 | 0.9868 | 0.3709 | 8,826 | |
| LWB | 1.0895 | 1.0708 | 1.3471 | 1.1552 | 1.1563 | 0.7096 | 0.4159 | 0.4855 | |
表5 基于13个基因座的9个品种间Nei氏无偏遗传距离(对角线下方)和遗传分化时间(年, 对角线上方)
Table 5 Nei′s unbiased genetic distance (below the diagonal) and genetic divergence time (years, above the diagonal) among nine breeds based on 13 loci
| 品种 Breeds | 品种 Breeds | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| STH | LTH | WD | SD | CB | JNG | LBW | LSM | LWB | |
| STH | 4,569 | 3,972 | 5,725 | 5,396 | 27,154 | 23,441 | 23,105 | 19,807 | |
| LTH | 0.2513 | 5,229 | 6,174 | 8,405 | 25,063 | 24,943 | 25,639 | 19,467 | |
| WD | 0.2185 | 0.2876 | 5,092 | 8,894 | 28,713 | 22,550 | 24,521 | 24,490 | |
| SD | 0.3149 | 0.3396 | 0.2801 | 9,286 | 28,955 | 24,108 | 25,823 | 21,002 | |
| CB | 0.2968 | 0.4623 | 0.4892 | 0.5108 | 34,493 | 26,606 | 30,001 | 21,022 | |
| JNG | 1.4936 | 1.3786 | 1.5794 | 1.5927 | 1.8973 | 10,626 | 17,940 | 12,901 | |
| LBW | 1.2894 | 1.3720 | 1.2404 | 1.3261 | 1.4635 | 0.5845 | 6,743 | 7,561 | |
| LSM | 1.2709 | 1.4103 | 1.3488 | 1.4204 | 1.6502 | 0.9868 | 0.3709 | 8,826 | |
| LWB | 1.0895 | 1.0708 | 1.3471 | 1.1552 | 1.1563 | 0.7096 | 0.4159 | 0.4855 | |
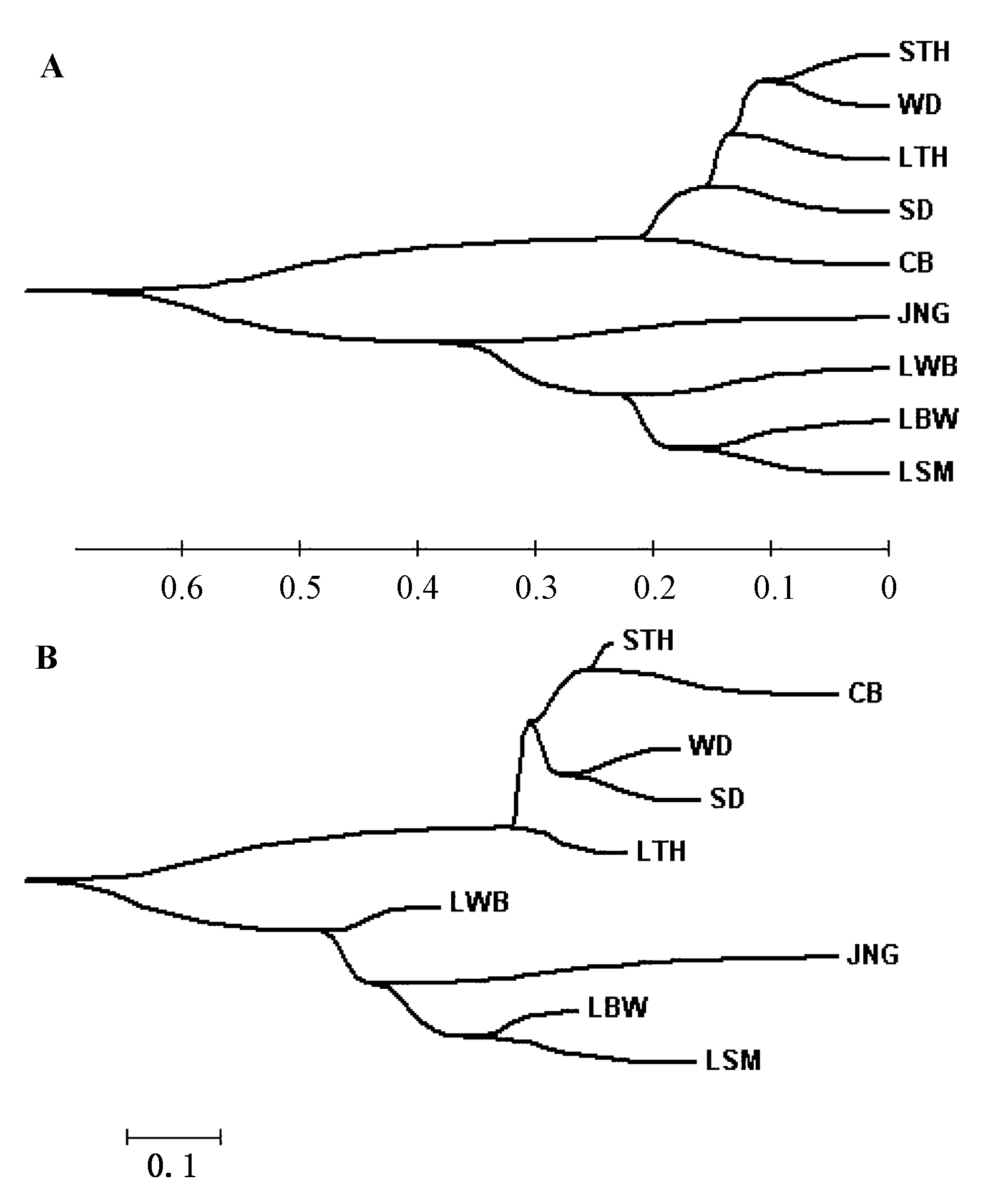
图1 基于Nei’s无偏遗传距离的9个品种的UPGMA(A)和ME(B)系统发生树。树图系由MEGA3.1软件绘制。品种代号同表1。
Fig. 1 UPGMA (A) and ME (B) phylogenetic dendrograms based on Nei's unbiased genetic distance of nine breeds. Dendrograms are drawn using MEGA 3.1 software. Breed codes are given in Table 1.
| [1] | Allard MW, Miyamoto MM, Jarecki L, Kraus F, Tennant MR (1992) DNA systematics and evolution of the artiodactyl family Bovidae. Proceedings of the National Academy of Sciences,USA, 89,3972-3976. |
| [2] | Arranz JJ, Bayon Y (2001) Genetic variation at microsatellite loci in Spanish sheep. Small Ruminant Research, 39,3-10. |
| [3] | Balloux F, Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Molecular Ecology, 11,155-165. |
| [4] |
Baumung R, Cubric-Curik V, Schwend K, Achmann R, Sölkne J (2006) Genetic characterization and breed assignment in Austrian sheep breeds using microsatellite marker information. Journal of Animal Breeding and Genetics, 123,265-277.
URL PMID |
| [5] | Bishop MD, Kappes SM, Keele JW, Stone RT, Sunden SLF, Hawkins GA, Toldo SS, Fries R, Grosz MD, Yoo J, Beattie CW (1994) A genetic linkage map for cattle. Genetics, 136,619-639. |
| [6] | Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment polymorphisms. American Journal of Human Genetics, 32,314-331. |
| [7] | Braend M, Tucker EM (1988) Hemoglobin types in Saanen goats and Barbary sheep: genetic and comparative aspects. Biochemical Genetics, 26,511-518. |
| [8] | Buchanan FC, Adams LJ (1994) Determination of evolutionary relationships among sheep breeds using microsatellites. Genomics, 22,397-403. |
| [9] | Cañón J, García D, García-Atance MA, Obexer-Ruff G, Lenstra JA, Ajmone-Marsan P, Dunner S (2006) Geographical partitioning of goat diversity in Europe and the Middle East. Animal Genetics, 37,327-336. |
| [10] | Cao LR (曹丽荣), Wang XM (王小明), Rao G (饶刚), Wan QH (万秋红), Fang SG (方盛国) (2004) The phylogenetic relationship among goat, sheep and bharal based on mitochondrial cytochrome b gene sequences . Acta Theriolo- gica Sinica (兽类学报), 24,109-114. (in Chinese with English abstract) |
| [11] | Cargill SH, Anderson GB, Medrano JF (1995) Development of a specific marker using RAPD analysis to distinguish between sheep and goats. Animal Biotechnology, 6,93-100. |
| [12] | Chang H (常洪) (1998) Studies on Animal Genetic Resources in China (中国家畜遗传资源研究). Shaanxi People’s Education Press, Xi’an. (in Chinese) |
| [13] | Chen SY (陈善元), Zhang YP (张亚平) (2006) Genetic approaches to study origins of domestic animals. Chinese Science Bulletin (科学通报), 51,2469-2475. (in Chinese) |
| [14] | Chen YC (陈幼春), Cao HH (曹红鹤), Li HB (李宏滨) (2001) Application of random-locating sampling in subpopulation of animal breed. Journal of Yellow Cattle Science (黄牛杂志), 27(1),1-3. (in Chinese with English abstract) |
| [15] | Chen YC (陈幼春), Ma YH (马月辉), Wang DY (王端云) (2003) Elements and achievement in research on domestic animal biodiversity. Biodiversity Science (生物多样性), 11,407-413. (in Chinese with English abstract) |
| [16] | Chen YC (陈幼春), Ma YH (马月辉), Pu YB (浦亚斌), Lv SJ (吕慎金) (2005) Relationship of selection of domestic animals and caused gene frequencies variation with cluster analysis and breed conservation. Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 36,1236-1240. (in Chinese with English abstract) |
| [17] | Crawford AM, Cuthbertson RP (1996) Mutations in sheep microsatellites. Genome Research, 6,876-879. |
| [18] | de Gortari MJ, Freking BA, Cuthbertson RP, Kappes SM, Keele JW, Stone RT, Leymaster KA, Dodds KG, Crawford AM, Beattie CW (1998) A second-generation linkage map of the sheep genome. Mammalian Genome, 9,204-209. |
| [19] | FAO (1998) Secondary Guidelines for the Development of National Farm Animal Genetic Resources Management Plans: Management of Small Populations at Risk, FAO, UNEP, Rome. |
| [20] | Fitch WM, Langley CH (1976) Protein evolution and the molecular clock. Federation Proceedings, 35, 2092-2097. |
| [21] | Geist V (1971) Mountain Sheep: A Study in Behavior and Evolution, pp.120-126. University of Chicago Press, Chicago. |
| [22] | Goudet J (2001) FSTAT, A Program to Stimate and Test Gene Diversities and Fixation Indices. Version 2.9.3 http://www2unil.ch/popgen/softwares/fstat.htm. |
| [23] | Gu WC (顾万春) (2004) Statistical Genetics (统计遗传学). Science Press, Beijing. (in Chinese) |
| [24] | Hammer S, Nadlinger K, Hartl GB (1995) Mitochondrial-DNA differentiation in chamois (genus Rupicapra): implications for taxonomy, conservation, and management. Acta Theriologica, 3 (Suppl.),145-155. |
| [25] | Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics, 5,150-163. |
| [26] | Li XL (李祥龙), Zheng GR (郑桂茹), Zhang YP (张亚平) (2000) Study on the genetic differentiation among sheep, goat and bharal based on the restriction fragment length polymorphism of mtDNA. Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 31,289-295. (in Chinese with English abstract) |
| [27] | Li XL, Alessio V (2004) Genetic diversity of Chinese indigenous goat breeds based on microsatellite markers. Journal of Animal Breeding and Genetics, 121,350-359. |
| [28] | Luo YF (罗永发), Wang ZG (王志刚), Li JQ (李加琪), Zhang GX (张桂香), Chen YS (陈瑶生), Liang Y (梁勇), Yu FQ (于福清), Song WT (宋卫涛), Zhang ZF (张自富) (2006) Genetic variation and genetic relationship among 13 Chinese and introduced cattle breeds using microsatellite DNA markers. Biodiversity Science (生物多样性), 14,498-507. (in Chinese with English abstract) |
| [29] | Ma YH, Rao SQ, Lu SJ, Hou GY, Guan WJ, Li HB, Li X, Zhao QJ, Guo J (2006) Phylogeography and origin of sheep breeds in northern China. Conservation Genetics, 7,117-127. |
| [30] | Manly BFJ (1985) The Statistics of Natural Selection. pp.272-282. Chapman and Hall, London. |
| [31] | Mattapallil MJ, Ali S (1999) Analysis of conserved microsatellite sequences suggests closer relationship between water buffalo Bubalus bubalis and sheep Ovis aries . DNA and Cell Biology, 18,513-519. |
| [32] | Meng SJ, Wang PZ (1997) Mitochondrial DNA polymorphisms for takin, cattle and sheep in Bovidae. In:Proceedings of International Conference on Animal Biotechnology, pp.174-177. International Academic Publishers, Beijing. |
| [33] | Metehan U, Beatriz GG, Juan-José A, Fermín SP, Mustafa S, Mehmet K, Yolanda B (2006) Genetic relationships among Turkish sheep. Genetics Selection Evolution, 38,513-524. |
| [34] |
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89,583-590.
URL PMID |
| [35] |
Pérez T, Albornoz J, Domínguez A (2002) Phylogeography of chamois ( Rupicapra spp.) inferred from microsatellites . Molecular Phylogenetics and Evolution, 25,524-534.
DOI URL PMID |
| [36] | Qi JF (齐景发) (2004) Report on Domestic Animal Genetic Resources in China (中国畜禽遗传资源状况). China Agriculture Press, Beijing. (in Chinese) |
| [37] | Saitbekova N, Gaillard C, Obexer-Ruff G, Dolf G (1999) Genetic diversity in Swiss goat breeds based on microsatellite analysis. Animal Genetics, 30,36-41. |
| [38] | Sambrook J, Fritsch FE, Mariatis T, (translated by Jin DY(金冬雁), Li MF (黎孟枫), Hou YD (侯云德) (1998) Molecular Cloning: A Laboratory Manual, 2nd edn, pp.304-315. Science Press, Beijing. (in Chinese) |
| [39] | Si JC (司俊臣) (1999) Breeds of Domestic Animal and Poultry in Shandong Province (山东省畜禽品种志), pp.51-59. Seasky Publishing House, Shenzhen. (in Chinese) |
| [40] |
Stone RT, Pulido JC, Duyk GM, Kappes SM, Keele JW, Beattie CW (1995) A small-insert bovine genomic library highly enriched for microsatellite repeat sequences. Mammalian Genome, 6,714-724.
DOI URL PMID |
| [41] | Sun YD (孙允东), Ma YH (马月辉), Wang JM (王建民), Li HB (李宏滨), Yuan CZ (苑存忠), Shang YG (尚友国) (2006) Genetic diversity of microsatellite loci in indigenous goat breeds in Shandong Province. Acta Ecologiae Animalis Domastici (家畜生态学报), 27,19-27. (in Chinese with English abstract) |
| [42] | Tomiuk J, Guldbrandtsen B, Loeschcke V (1998) Population differentiation through mutation and drift—a comparison of genetic identity measures. Genetica, 102/103,545-558. |
| [43] | Tu YR (涂友仁) (1989) Sheep and Goat Breeds in China (中国羊品种志). Shanghai Science & Technology Publishers, Shanghai. (in Chinese) |
| [44] | Weir BS translated by Xu YB (徐云碧), Wang ZN (王志宁), Yu ZH (俞志华), (1996) Genetic Data Analysis, pp.152-155. China Agriculture Press, Beijing. (in Chinese) |
| [45] | Yan LN (闫路娜), Zhang DX (张德兴) (2004) Effects of sample size on various genetic diversity measures in population genetic study with microsatellite DNA markers. Acta Zoologica Sinica (动物学报), 50,279-290. (in Chinese with English abstract) |
| [46] | Yang Y (杨燕), Ma YH (马月辉), Lu SJ (吕慎金), Zhang YH (张英汉) (2004) Genetic diversity in seven Chinese indigenous sheep breeds based on microsatellite analysis. Biodiversity Science (生物多样性), 12,586-593. (in Chinese with English abstract) |
| [47] | Yang ZP (杨章平), Chang H (常洪), Sun W (孙伟), Geng RQ (耿荣庆), Mao YJ (毛永江) (2003) The comparison of structural loci and microsatellite marker on genetic differentiation in sheep (goat) populations. Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 34,427-433. (in Chinese with English abstract) |
| [48] | Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian Journal of Botany, 129,157. |
| [49] | Yuan CZ (苑存忠), Wang JM (王建民), Ma YH (马月辉), Qu XX (曲绪仙), Shang YG (尚友国), Zhang NB (张宁波) (2006) Genetic diversity of indigenous sheep breeds in Shandong Province based on microsatellite markers study. Chinese Journal of Applied Ecology (应用生态学报), 17,1459-1464. (in Chinese with English abstract) |
| [50] | Zhang CS (张传生), Geng LY (耿立英), Wan HW (万海伟), Li JT (李吉涛), Du LX (杜立新) (2005) Study on sequence variation of mitochondrial cytochrome b gene and phylogenetic relationship of four native sheep breeds . Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 36,313-317. (in Chinese with English abstract) |
| [1] | 张静, 李渊, 宋娜, 林龙山, 高天翔. 我国沿海棱鳀属鱼类的物种鉴定与系统发育[J]. 生物多样性, 2016, 24(8): 888-895. |
| [2] | 钱贞娜, 孟千万, 任明迅. 风筝果镜像花的雌雄异位变化及传粉生态型的形成[J]. 生物多样性, 2016, 24(12): 1364-1372. |
| [3] | 李春楠, 崔海瑞, 王伟博. 用SRAP标记研究根际土壤微生物的遗传多样性[J]. 生物多样性, 2011, 19(4): 485-493. |
| [4] | 张俊红, 黄华宏, 童再康, 程龙军, 梁跃龙, 陈奕良. 光皮桦6个南方天然群体的遗传多样性[J]. 生物多样性, 2010, 18(3): 233-240. |
| [5] | 戴李川, 张明龙, 刘继业, 李小白, 崔海瑞. 用EST-SSR标记检测我国甘蓝型油菜品种的遗传多样性[J]. 生物多样性, 2009, 17(5): 482-489. |
| [6] | 孟彦, 杨焱清, 张燕, 肖汉兵. 野生和养殖大鲵群体遗传多样性的微卫星分析[J]. 生物多样性, 2008, 16(6): 533-538. |
| [7] | 魏麟, 刘胜贵, 史宪伟. 雪峰乌骨鸡自然群体遗传多样性的微卫星分析[J]. 生物多样性, 2008, 16(5): 503-508. |
| [8] | 赵晓枫, 吴俊红, 徐宁迎, 胡晓湘, 李宁. 金华猪遗传结构及其与太湖猪遗传分化的研究[J]. 生物多样性, 2008, 16(4): 339-345. |
| [9] | 杨燕, 马月辉, 吕慎金, 张英汉. 中国7个地方绵羊品种遗传多样性的微卫星分析[J]. 生物多样性, 2004, 12(6): 586-593. |
| [10] | 汪小凡, 廖万金, 宋志平. 小毛茛居群的遗传分化及其与空间隔离的相关性[J]. 生物多样性, 2001, 09(2): 138-144. |
| [11] | 胡文平, 连林生, 宿兵, 聂龙, 张亚平. 滇南小耳猪遗传多样性的血液蛋白电泳研究[J]. 生物多样性, 1998, 06(1): 22-26. |
| [12] | 郑向忠, 徐宏发, 陆厚基. 动物种群遗传异质性研究进展*[J]. 生物多样性, 1997, 05(3): 210-216. |
| [13] | 聂龙, 施立明. 西南地区地方品种猪血液蛋白遗传多样性研究[J]. 生物多样性, 1995, 03(1): 1-7. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn