



Biodiv Sci ›› 2018, Vol. 26 ›› Issue (3): 217-228. DOI: 10.17520/biods.2017336 cstr: 32101.14.biods.2017336
Special Issue: 物种形成与系统进化
• Original Papers: Plant Diversity • Next Articles
Hou Qinxi1,2, Ci Xiuqin1,2, Liu Zhifang1,2, Xu Wumei3, Li Jie1,*( )
)
Received:2017-12-25
Accepted:2018-02-24
Online:2018-03-20
Published:2018-05-05
Contact:
Li Jie
Hou Qinxi, Ci Xiuqin, Liu Zhifang, Xu Wumei, Li Jie. Assessment of the evolutionary history of Lauraceae in Xishuangbanna National Nature Reserve using DNA barcoding[J]. Biodiv Sci, 2018, 26(3): 217-228.
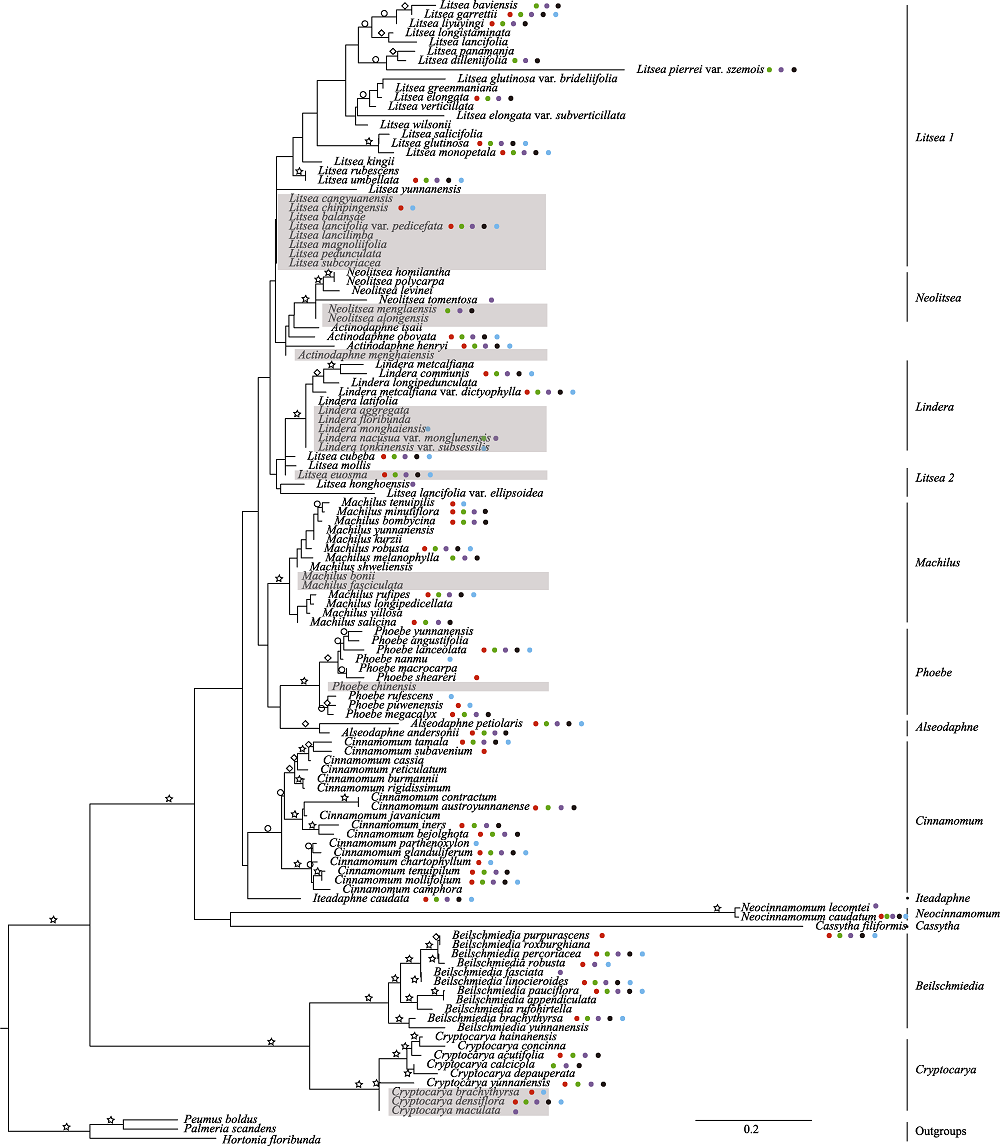
Fig. 1 A phylogenetic tree of 121 species of Lauraceae in Xishuangbanna based on a maximum likelihood analysis of ITS sequence data. Nodes with strong (≥ 85%), moderate (70-84%) and weak (50-69%) bootstrap support are indicated by an asterisk, a diamond and a round, respectively. The 23 species without molecular information were highlighted with gray bars. Round filled with red, green, purple, black, blue represent the distribution of the species in Mengyang sub-reserve, Menglun sub-reserve, Mengla sub-reserve, Shangyong sub-reserve, and Mangao sub-reserve, respectively.
| 区域 Region | 物种丰富度 SR (%) | 系统发育多样性 PDFaith (%) | 标准化系统发育多样性 Standardized effect size of PD (PDSES) | P |
|---|---|---|---|---|
| 西双版纳国家级自然保护区 Xishuangbanna National Nature Reserve | 66 (54.5) | 5.78 (88.8) | - | - |
| 勐养子保护区 Mengyang sub-reserve | 49 (40.5) | 5.41(83.1) | 1.15 | 0.85 |
| 勐仑子保护区 Menglun sub-reserve | 47 (38.8) | 5.49 (84.3) | 1.35 | 0.89 |
| 勐腊子保护区 Mengla sub-reserve | 53 (43.8) | 5.64 (86.6) | 1.21 | 0.88 |
| 尚勇子保护区 Shangyong sub-reserve | 47 (38.8) | 5.49 (84.3) | 1.32 | 0.89 |
| 曼稿子保护区 Mangao sub-reserve | 36 (29.8) | 4.97 (76.3) | 1.54 | 0.92 |
| 西双版纳地区(总) Xishuangbanna region (Total) | 121 | 6.51 | - | - |
Table 1 Phylogenetic diversity (PDFaith), species richness (SR) and their proportions of Lauraceae in Xishuangbanna National Nature Reserve and each sub-reserve
| 区域 Region | 物种丰富度 SR (%) | 系统发育多样性 PDFaith (%) | 标准化系统发育多样性 Standardized effect size of PD (PDSES) | P |
|---|---|---|---|---|
| 西双版纳国家级自然保护区 Xishuangbanna National Nature Reserve | 66 (54.5) | 5.78 (88.8) | - | - |
| 勐养子保护区 Mengyang sub-reserve | 49 (40.5) | 5.41(83.1) | 1.15 | 0.85 |
| 勐仑子保护区 Menglun sub-reserve | 47 (38.8) | 5.49 (84.3) | 1.35 | 0.89 |
| 勐腊子保护区 Mengla sub-reserve | 53 (43.8) | 5.64 (86.6) | 1.21 | 0.88 |
| 尚勇子保护区 Shangyong sub-reserve | 47 (38.8) | 5.49 (84.3) | 1.32 | 0.89 |
| 曼稿子保护区 Mangao sub-reserve | 36 (29.8) | 4.97 (76.3) | 1.54 | 0.92 |
| 西双版纳地区(总) Xishuangbanna region (Total) | 121 | 6.51 | - | - |
| 西双版纳行政乡镇 Administrative towns in Xishuangbanna | SR | PDFaith | PDSES | P | |
|---|---|---|---|---|---|
| 勐海县 Menghai County | 勐满镇 Mengman Town | 39 | 4.96 | 1.16 | 0.87 |
| 勐阿镇 Menga Town | 24 | 3.72 | 1.00 | 0.82 | |
| 勐往乡 Mengwang Town | 26 | 4.63 | 2.06 | 0.97 | |
| 西定布朗族哈尼族乡 Xiding Blang Hani Ethnicity Town | 17 | 2.00 | -0.91 | 0.17 | |
| 勐遮镇 Mengzhe Town | 20 | 3.13 | 0.51 | 0.70 | |
| 勐海镇 Menghai Town | 12 | 1.24 | -1.46 | 0.06 | |
| 勐宋乡 Mengsong Town | 26 | 2.27 | -1.49 | 0.04 | |
| 打洛镇 Daluo Town | 30 | 4.83 | 1.95 | 0.96 | |
| 勐混镇 Menghun Town | 31 | 4.64 | 1.57 | 0.93 | |
| 格朗和哈尼族乡 Gelanghe Hani Ethnicity Town | 45 | 4.10 | -0.43 | 0.35 | |
| 布朗山布朗族乡 Bulangshan Blang Ethnicity Town | 13 | 2.12 | -0.08 | 0.55 | |
| 景洪市 Jinghong City | 景讷乡 Jingne Town | 29 | 3.64 | 0.25 | 0.59 |
| 普文镇 Puwen Town | 17 | 4.07 | 2.59 | 0.99 | |
| 勐旺乡 Mengwang Town | 21 | 3.62 | 1.16 | 0.87 | |
| 大渡岗乡 Dadugang Town | 22 | 3.35 | 0.62 | 0.74 | |
| 景洪 Jinghong | 14 | 2.29 | 0.02 | 0.58 | |
| 勐养镇 Mengyang Town | 38 | 4.92 | 1.20 | 0.87 | |
| 基诺山基诺族乡Jinuoshan Jinuo Ethnicity Town | 52 | 4.33 | -0.65 | 0.26 | |
| 嘎洒镇 Gasa Town | 20 | 2.71 | -0.11 | 0.53 | |
| 勐罕镇 Menghan Town | 16 | 1.78 | -1.13 | 0.07 | |
| 景哈哈尼族乡 Jingha Hani Ethnicity Town | 15 | 2.23 | -0.20 | 0.48 | |
| 勐龙镇 Menglong Town | 53 | 4.39 | -0.58 | 0.30 | |
| 勐腊县 Mengla County | 象明彝族乡 Xiangming Yi Ethnicity Town | 43 | 5.02 | 0.99 | 0.83 |
| 易武乡 Yiwu Town | 26 | 4.58 | 2.11 | 0.97 | |
| 勐仑镇 Menglun Town | 63 | 5.04 | -0.42 | 0.33 | |
| 关累镇 Guanlei Town | 28 | 2.74 | -0.98 | 0.17 | |
| 瑶区瑶族乡 Yaoqu Yao Ethnicity Town | 14 | 2.10 | -0.28 | 0.47 | |
| 勐伴镇 Mengban Town | 38 | 4.00 | -0.03 | 0.50 | |
| 勐捧镇 Mengpeng Town | 15 | 2.15 | -0.30 | 0.46 | |
| 勐腊镇 Mengla Town | 66 | 4.92 | -0.76 | 0.21 | |
| 勐满镇 Mengman Town | 15 | 1.73 | -1.09 | 0.08 | |
| 尚勇镇 Shangyong Town | 29 | 4.62 | 1.77 | 0.95 | |
Table 2 Species richness (SR), phylogenetic diversity (PDFaith) and the standardized effect size of PD (PDSES) of Lauraceae in 32 administrative towns in Xishuangbanna
| 西双版纳行政乡镇 Administrative towns in Xishuangbanna | SR | PDFaith | PDSES | P | |
|---|---|---|---|---|---|
| 勐海县 Menghai County | 勐满镇 Mengman Town | 39 | 4.96 | 1.16 | 0.87 |
| 勐阿镇 Menga Town | 24 | 3.72 | 1.00 | 0.82 | |
| 勐往乡 Mengwang Town | 26 | 4.63 | 2.06 | 0.97 | |
| 西定布朗族哈尼族乡 Xiding Blang Hani Ethnicity Town | 17 | 2.00 | -0.91 | 0.17 | |
| 勐遮镇 Mengzhe Town | 20 | 3.13 | 0.51 | 0.70 | |
| 勐海镇 Menghai Town | 12 | 1.24 | -1.46 | 0.06 | |
| 勐宋乡 Mengsong Town | 26 | 2.27 | -1.49 | 0.04 | |
| 打洛镇 Daluo Town | 30 | 4.83 | 1.95 | 0.96 | |
| 勐混镇 Menghun Town | 31 | 4.64 | 1.57 | 0.93 | |
| 格朗和哈尼族乡 Gelanghe Hani Ethnicity Town | 45 | 4.10 | -0.43 | 0.35 | |
| 布朗山布朗族乡 Bulangshan Blang Ethnicity Town | 13 | 2.12 | -0.08 | 0.55 | |
| 景洪市 Jinghong City | 景讷乡 Jingne Town | 29 | 3.64 | 0.25 | 0.59 |
| 普文镇 Puwen Town | 17 | 4.07 | 2.59 | 0.99 | |
| 勐旺乡 Mengwang Town | 21 | 3.62 | 1.16 | 0.87 | |
| 大渡岗乡 Dadugang Town | 22 | 3.35 | 0.62 | 0.74 | |
| 景洪 Jinghong | 14 | 2.29 | 0.02 | 0.58 | |
| 勐养镇 Mengyang Town | 38 | 4.92 | 1.20 | 0.87 | |
| 基诺山基诺族乡Jinuoshan Jinuo Ethnicity Town | 52 | 4.33 | -0.65 | 0.26 | |
| 嘎洒镇 Gasa Town | 20 | 2.71 | -0.11 | 0.53 | |
| 勐罕镇 Menghan Town | 16 | 1.78 | -1.13 | 0.07 | |
| 景哈哈尼族乡 Jingha Hani Ethnicity Town | 15 | 2.23 | -0.20 | 0.48 | |
| 勐龙镇 Menglong Town | 53 | 4.39 | -0.58 | 0.30 | |
| 勐腊县 Mengla County | 象明彝族乡 Xiangming Yi Ethnicity Town | 43 | 5.02 | 0.99 | 0.83 |
| 易武乡 Yiwu Town | 26 | 4.58 | 2.11 | 0.97 | |
| 勐仑镇 Menglun Town | 63 | 5.04 | -0.42 | 0.33 | |
| 关累镇 Guanlei Town | 28 | 2.74 | -0.98 | 0.17 | |
| 瑶区瑶族乡 Yaoqu Yao Ethnicity Town | 14 | 2.10 | -0.28 | 0.47 | |
| 勐伴镇 Mengban Town | 38 | 4.00 | -0.03 | 0.50 | |
| 勐捧镇 Mengpeng Town | 15 | 2.15 | -0.30 | 0.46 | |
| 勐腊镇 Mengla Town | 66 | 4.92 | -0.76 | 0.21 | |
| 勐满镇 Mengman Town | 15 | 1.73 | -1.09 | 0.08 | |
| 尚勇镇 Shangyong Town | 29 | 4.62 | 1.77 | 0.95 | |
| 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution | 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution |
|---|---|---|---|---|---|---|---|---|---|
| 思茅黄肉楠 Actinodaphne henryi | 0.11 | EN | - | + | 无根藤 Cassytha filiformis | 1.08 | - | VU | + |
| 勐海黄肉楠 A. menghaiensis | 0.02 | - | VU | - | 滇南桂 Cinnamomum austro-yunnanense | 0.06 | VU | VU | + |
| 倒卵叶黄肉楠 A. obovata | 0.06 | - | - | + | 钝叶桂 C. bejolghota | 0.06 | - | - | + |
| 马关黄肉楠 A. tsaii | 0.05 | - | CR | - | 阴香 C. burmannii | 0.02 | - | - | - |
| 毛叶油丹 Alseodaphne andersonii | 0.07 | - | - | + | 肉桂 C. cassia | 0.02 | - | - | - |
| 长柄油丹 A. petiolaris | 0.19 | - | VU | + | 坚叶樟 C. chartophyllum | 0.04 | EN | VU | + |
| 山潺 Beilschmiedia appendiculata | 0.05 | - | - | - | 聚花桂 C. contractum | 0.06 | - | - | - |
| 勐仑琼楠 B. brachythyrsa | 0.12 | EN | VU | + | 云南樟 C. glanduliferum | 0.03 | - | - | + |
| 白柴果 B. fasciata | 0.03 | EN | - | + | 大叶桂 C. iners | 0.06 | - | - | + |
| 李榄琼楠 B. linocieroides | 0.03 | VU | - | + | 爪哇肉桂 C. javanicum | 0.02 | - | - | - |
| 少花琼楠 B. pauciflora | 0.06 | EN | VU | + | 毛叶樟 C. mollifolium | 0.02 | EN | VU | + |
| 厚叶琼楠 B. percoriacea | 0.03 | - | - | + | 黄樟 C. parthenoxylon | 0.02 | - | - | + |
| 紫叶琼楠 B. purpurascens | 0.03 | EN | VU | + | 网脉桂 C. reticulatum | 0.03 | - | - | - |
| 粗壮琼楠 B. robusta | 0.04 | - | - | + | 卵叶桂 C. rigidissimum | 0.02 | - | VU | - |
| 椆琼楠 B. roxburghiana | 0.03 | - | - | - | 香桂 C. subavenium | 0.04 | - | VU | + |
| 红毛琼楠 B. rufohirtella | 0.09 | EN | VU | - | 柴桂 C. tamala | 0.04 | - | - | + |
| 滇琼楠 B. yunnanensis | 0.17 | - | - | - | 细毛樟 C. tenuipilum | 0.03 | - | - | + |
| 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution | 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution |
| 樟 C. camphora | 0.06 | - | - | - | 假柿木姜子 L. monopetala | 0.09 | - | - | + |
| 尖叶厚壳桂 Cryptocarya acutifolia | 0.08 | - | - | + | 香花木姜子 L. panamonja | 0.06 | - | - | - |
| 短序厚壳桂 C. brachythyrsa | 0.17 | EN | - | + | 红皮木姜子 L. pedunculata | 0.01 | - | - | - |
| 岩生厚壳桂 C. calcicola | 0.06 | - | - | + | 思茅木姜子 L. pierrei var. szemois | 0.45 | EN | VU | + |
| 黄果厚壳桂 C. concinna | 0.08 | - | - | - | 红叶木姜子 L. rubescens | 0.02 | - | - | - |
| 丛花厚壳桂 C. densiflora | 0.17 | - | - | + | 黑木姜子 L. salicifolia | 0.05 | - | - | - |
| 贫花厚壳桂 C. depauperata | 0.10 | - | - | - | 桂北木姜子 L. subcoriacea | 0.01 | - | - | - |
| 海南厚壳桂 C. hainanensis | 0.04 | - | EN | - | 伞花木姜子 L. umbellate | 0.02 | - | - | + |
| 斑果厚壳桂 C. maculata | 0.17 | CR | - | + | 轮叶木姜子 L. verticillata | 0.03 | - | - | - |
| 云南厚壳桂 C. yunnanensis | 0.23 | EN | - | + | 绒叶木姜子 L. wilsonii | 0.05 | - | - | - |
| 香面叶 Iteadaphne caudata | 0.15 | - | - | + | 云南木姜子 L. yunnanensis | 0.15 | - | VU | - |
| 乌药 Lindera aggregata | 0.02 | - | - | - | 长梗润楠 Machilus longipedicellata | 0.02 | - | - | - |
| 香叶树 L. communis | 0.07 | - | - | + | 黄心树 M. bombycina | 0.01 | - | - | + |
| 绒毛钓樟 L. floribunda | 0.02 | - | VU | - | 枇杷叶润楠 M. bonii | 0.04 | - | - | - |
| 团香果 L. latifolia | 0.02 | - | - | - | 簇序润楠 M. fasciculata | 0.04 | CR | - | - |
| 山柿子果 L. longipedunculata | 0.04 | VU | - | - | 秃枝润楠 M. kurzii | 0.01 | - | VU | - |
| 滇粤山胡椒 L. metcalfiana | 0.06 | - | - | - | 暗叶润楠 M. melanophylla | 0.04 | CR | VU | + |
| 网叶山胡椒 L. metcalfiana var. dictyophylla | 0.04 | - | - | + | 小花润楠 M. minutiflora | 0.01 | EN | VU | + |
| 勐海山胡椒 L. monghaiensis | 0.02 | CR | VU | + | 粗壮润楠 M. robusta | 0.02 | - | - | + |
| 勐仑山胡椒 L. nacusua var. monglunensis | 0.02 | - | VU | + | 红梗润楠 M. rufipes | 0.02 | - | - | + |
| 无梗假桂钓樟 L. tonkinensis var. subsessilis | 0.02 | - | VU | + | 柳叶润楠 M. salicina | 0.03 | - | - | + |
| 假辣子 Litsea balansae | 0.01 | VU | - | - | 瑞丽润楠 M. shweliensis | 0.03 | VU | - | - |
| 大萼木姜子 L. baviensis | 0.07 | - | VU | + | 细毛润楠 M. tenuipila | 0.02 | - | VU | + |
| 沧原木姜子 L. cangyuanensis | 0.01 | - | - | - | 柔毛润楠 M. villosa | 0.03 | - | - | - |
| 金平木姜子 L. chinpingensis | 0.01 | - | VU | + | 滇润楠 M. yunnanensis | 0.01 | - | - | - |
| 山鸡椒 L. cubeba | 0.03 | - | - | + | 滇新樟 Neocinnamomum caudatum | 0.49 | - | - | + |
| 五桠果叶木姜子 L. dilleniifolia | 0.04 | EN | VU | + | 海南新樟 N. lecomtei | 0.49 | - | VU | + |
| 黄丹木姜子 L. elongata | 0.02 | - | - | + | 下龙新木姜子 Neolisea alongensis | 0.02 | - | CR | - |
| 近轮叶木姜子 L. elongata var. subverticillata | 0.16 | - | - | - | 团花新木姜子 N. homilantha | 0.02 | - | - | - |
| 清香木姜子 L. euosma | 0.02 | - | - | + | 大叶新木姜子 N. levinei | 0.05 | - | VU | - |
| 滇南木姜子 L. garrettii | 0.03 | - | - | + | 勐腊新木姜子 N. menglaensis | 0.02 | CR | VU | + |
| 潺槁木姜子 L. glutinosa | 0.03 | - | - | + | 多果新木姜子 N. polycarpa | 0.02 | - | - | - |
| 白野槁树 L. glutinosa var. bridellifolia | 0.12 | - | - | - | 绒毛新木姜子 N. tomentosa | 0.11 | EN | - | + |
| 华南木姜子 L. greenmaniana | 0.01 | - | - | - | 沼楠 Phoebe angustifolia | 0.03 | - | CR | - |
| 红河木姜子 L. honghoensis | 0.06 | EN | - | + | 山楠 P. chinensis | 0.05 | - | - | - |
| 秃净木姜子 L. kingii | 0.05 | - | - | - | 披针叶楠 P. lanceolata | 0.05 | - | - | + |
| 剑叶木姜子 L. lancifolia | 0.08 | - | - | - | 大果楠 P. macrocarpa | 0.03 | EN | VU | - |
| 椭圆果木姜子 L. lancifoiia var. ellipsoidea | 0.01 | - | - | - | 大萼楠 P. megacalyx | 0.04 | EN | VU | + |
| 有梗木姜子 L. lancifolia var. pedicefata | 0.18 | - | - | + | 滇楠 P. nanmu | 0.03 | EN | VU | + |
| 大果木姜子 L. lancilimba | 0.01 | - | VU | - | 普文楠 P. puwenensis | 0.03 | EN | - | + |
| 圆锥木姜子 L. liyuyingi | 0.03 | - | - | + | 红梗楠 P. rufescens | 0.03 | EN | VU | + |
| 长蕊木姜子 L. longistaminata | 0.03 | VU | - | - | 紫楠 P. sheareri | 0.06 | - | - | + |
| 玉兰叶木姜子 L. magnoliifolia | 0.01 | - | VU | - | 景东楠 P. yunnanensis | 0.03 | EN | VU | - |
| 毛叶木姜子 L. mollis | 0.04 | - | - | - |
Table 3 Evolutionary distinctiveness (ED) and endangered levels (based on China Species Red List (I) and the result of Endemic Plants Inventory and Protection in Southwest China (II)) of Lauraceae in Xishuangbanna and distribution in the nature reserve
| 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution | 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution |
|---|---|---|---|---|---|---|---|---|---|
| 思茅黄肉楠 Actinodaphne henryi | 0.11 | EN | - | + | 无根藤 Cassytha filiformis | 1.08 | - | VU | + |
| 勐海黄肉楠 A. menghaiensis | 0.02 | - | VU | - | 滇南桂 Cinnamomum austro-yunnanense | 0.06 | VU | VU | + |
| 倒卵叶黄肉楠 A. obovata | 0.06 | - | - | + | 钝叶桂 C. bejolghota | 0.06 | - | - | + |
| 马关黄肉楠 A. tsaii | 0.05 | - | CR | - | 阴香 C. burmannii | 0.02 | - | - | - |
| 毛叶油丹 Alseodaphne andersonii | 0.07 | - | - | + | 肉桂 C. cassia | 0.02 | - | - | - |
| 长柄油丹 A. petiolaris | 0.19 | - | VU | + | 坚叶樟 C. chartophyllum | 0.04 | EN | VU | + |
| 山潺 Beilschmiedia appendiculata | 0.05 | - | - | - | 聚花桂 C. contractum | 0.06 | - | - | - |
| 勐仑琼楠 B. brachythyrsa | 0.12 | EN | VU | + | 云南樟 C. glanduliferum | 0.03 | - | - | + |
| 白柴果 B. fasciata | 0.03 | EN | - | + | 大叶桂 C. iners | 0.06 | - | - | + |
| 李榄琼楠 B. linocieroides | 0.03 | VU | - | + | 爪哇肉桂 C. javanicum | 0.02 | - | - | - |
| 少花琼楠 B. pauciflora | 0.06 | EN | VU | + | 毛叶樟 C. mollifolium | 0.02 | EN | VU | + |
| 厚叶琼楠 B. percoriacea | 0.03 | - | - | + | 黄樟 C. parthenoxylon | 0.02 | - | - | + |
| 紫叶琼楠 B. purpurascens | 0.03 | EN | VU | + | 网脉桂 C. reticulatum | 0.03 | - | - | - |
| 粗壮琼楠 B. robusta | 0.04 | - | - | + | 卵叶桂 C. rigidissimum | 0.02 | - | VU | - |
| 椆琼楠 B. roxburghiana | 0.03 | - | - | - | 香桂 C. subavenium | 0.04 | - | VU | + |
| 红毛琼楠 B. rufohirtella | 0.09 | EN | VU | - | 柴桂 C. tamala | 0.04 | - | - | + |
| 滇琼楠 B. yunnanensis | 0.17 | - | - | - | 细毛樟 C. tenuipilum | 0.03 | - | - | + |
| 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution | 物种 Species | ED | Ⅰ | Ⅱ | 分布 Distribution |
| 樟 C. camphora | 0.06 | - | - | - | 假柿木姜子 L. monopetala | 0.09 | - | - | + |
| 尖叶厚壳桂 Cryptocarya acutifolia | 0.08 | - | - | + | 香花木姜子 L. panamonja | 0.06 | - | - | - |
| 短序厚壳桂 C. brachythyrsa | 0.17 | EN | - | + | 红皮木姜子 L. pedunculata | 0.01 | - | - | - |
| 岩生厚壳桂 C. calcicola | 0.06 | - | - | + | 思茅木姜子 L. pierrei var. szemois | 0.45 | EN | VU | + |
| 黄果厚壳桂 C. concinna | 0.08 | - | - | - | 红叶木姜子 L. rubescens | 0.02 | - | - | - |
| 丛花厚壳桂 C. densiflora | 0.17 | - | - | + | 黑木姜子 L. salicifolia | 0.05 | - | - | - |
| 贫花厚壳桂 C. depauperata | 0.10 | - | - | - | 桂北木姜子 L. subcoriacea | 0.01 | - | - | - |
| 海南厚壳桂 C. hainanensis | 0.04 | - | EN | - | 伞花木姜子 L. umbellate | 0.02 | - | - | + |
| 斑果厚壳桂 C. maculata | 0.17 | CR | - | + | 轮叶木姜子 L. verticillata | 0.03 | - | - | - |
| 云南厚壳桂 C. yunnanensis | 0.23 | EN | - | + | 绒叶木姜子 L. wilsonii | 0.05 | - | - | - |
| 香面叶 Iteadaphne caudata | 0.15 | - | - | + | 云南木姜子 L. yunnanensis | 0.15 | - | VU | - |
| 乌药 Lindera aggregata | 0.02 | - | - | - | 长梗润楠 Machilus longipedicellata | 0.02 | - | - | - |
| 香叶树 L. communis | 0.07 | - | - | + | 黄心树 M. bombycina | 0.01 | - | - | + |
| 绒毛钓樟 L. floribunda | 0.02 | - | VU | - | 枇杷叶润楠 M. bonii | 0.04 | - | - | - |
| 团香果 L. latifolia | 0.02 | - | - | - | 簇序润楠 M. fasciculata | 0.04 | CR | - | - |
| 山柿子果 L. longipedunculata | 0.04 | VU | - | - | 秃枝润楠 M. kurzii | 0.01 | - | VU | - |
| 滇粤山胡椒 L. metcalfiana | 0.06 | - | - | - | 暗叶润楠 M. melanophylla | 0.04 | CR | VU | + |
| 网叶山胡椒 L. metcalfiana var. dictyophylla | 0.04 | - | - | + | 小花润楠 M. minutiflora | 0.01 | EN | VU | + |
| 勐海山胡椒 L. monghaiensis | 0.02 | CR | VU | + | 粗壮润楠 M. robusta | 0.02 | - | - | + |
| 勐仑山胡椒 L. nacusua var. monglunensis | 0.02 | - | VU | + | 红梗润楠 M. rufipes | 0.02 | - | - | + |
| 无梗假桂钓樟 L. tonkinensis var. subsessilis | 0.02 | - | VU | + | 柳叶润楠 M. salicina | 0.03 | - | - | + |
| 假辣子 Litsea balansae | 0.01 | VU | - | - | 瑞丽润楠 M. shweliensis | 0.03 | VU | - | - |
| 大萼木姜子 L. baviensis | 0.07 | - | VU | + | 细毛润楠 M. tenuipila | 0.02 | - | VU | + |
| 沧原木姜子 L. cangyuanensis | 0.01 | - | - | - | 柔毛润楠 M. villosa | 0.03 | - | - | - |
| 金平木姜子 L. chinpingensis | 0.01 | - | VU | + | 滇润楠 M. yunnanensis | 0.01 | - | - | - |
| 山鸡椒 L. cubeba | 0.03 | - | - | + | 滇新樟 Neocinnamomum caudatum | 0.49 | - | - | + |
| 五桠果叶木姜子 L. dilleniifolia | 0.04 | EN | VU | + | 海南新樟 N. lecomtei | 0.49 | - | VU | + |
| 黄丹木姜子 L. elongata | 0.02 | - | - | + | 下龙新木姜子 Neolisea alongensis | 0.02 | - | CR | - |
| 近轮叶木姜子 L. elongata var. subverticillata | 0.16 | - | - | - | 团花新木姜子 N. homilantha | 0.02 | - | - | - |
| 清香木姜子 L. euosma | 0.02 | - | - | + | 大叶新木姜子 N. levinei | 0.05 | - | VU | - |
| 滇南木姜子 L. garrettii | 0.03 | - | - | + | 勐腊新木姜子 N. menglaensis | 0.02 | CR | VU | + |
| 潺槁木姜子 L. glutinosa | 0.03 | - | - | + | 多果新木姜子 N. polycarpa | 0.02 | - | - | - |
| 白野槁树 L. glutinosa var. bridellifolia | 0.12 | - | - | - | 绒毛新木姜子 N. tomentosa | 0.11 | EN | - | + |
| 华南木姜子 L. greenmaniana | 0.01 | - | - | - | 沼楠 Phoebe angustifolia | 0.03 | - | CR | - |
| 红河木姜子 L. honghoensis | 0.06 | EN | - | + | 山楠 P. chinensis | 0.05 | - | - | - |
| 秃净木姜子 L. kingii | 0.05 | - | - | - | 披针叶楠 P. lanceolata | 0.05 | - | - | + |
| 剑叶木姜子 L. lancifolia | 0.08 | - | - | - | 大果楠 P. macrocarpa | 0.03 | EN | VU | - |
| 椭圆果木姜子 L. lancifoiia var. ellipsoidea | 0.01 | - | - | - | 大萼楠 P. megacalyx | 0.04 | EN | VU | + |
| 有梗木姜子 L. lancifolia var. pedicefata | 0.18 | - | - | + | 滇楠 P. nanmu | 0.03 | EN | VU | + |
| 大果木姜子 L. lancilimba | 0.01 | - | VU | - | 普文楠 P. puwenensis | 0.03 | EN | - | + |
| 圆锥木姜子 L. liyuyingi | 0.03 | - | - | + | 红梗楠 P. rufescens | 0.03 | EN | VU | + |
| 长蕊木姜子 L. longistaminata | 0.03 | VU | - | - | 紫楠 P. sheareri | 0.06 | - | - | + |
| 玉兰叶木姜子 L. magnoliifolia | 0.01 | - | VU | - | 景东楠 P. yunnanensis | 0.03 | EN | VU | - |
| 毛叶木姜子 L. mollis | 0.04 | - | - | - |
| [1] |
Abellán P, Sánchez-Fernández D, Picazo F, Millán A, Lobo JM, Ribera I (2013) Preserving the evolutionary history of freshwater biota in Iberian National Parks. Biological Conservation, 162, 116-126.
DOI URL |
| [2] |
Chen HF, Yi ZF, Schmidt-Vogt D, Ahrends A, Becksch?fer P, Kleinn C, Ranjitkar S, Xu JC (2016) Pushing the limits: The pattern and dynamics of rubber monoculture expansion in Xishuangbanna, SW China. PLoS ONE, 11, e0150062.
DOI URL PMID |
| [3] |
Chen S, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE, 5, e8613.
DOI URL PMID |
| [4] |
Ci XQ, Li J (2017) Phylogenetic diversity and its application in floristics and biodiversity conservation. Biodiversity Science, 25, 175-181. (in Chinese with English abstract)
DOI URL |
|
[慈秀芹, 李捷 (2017) 系统发育多样性在植物区系研究与生物多样性保护中的应用. 生物多样性, 25, 175-181.]
DOI URL |
|
| [5] | De Vos JM, Joppa LN, Gittleman JL, Stephens PR, Pimm SL (2015) Estimating the normal background rate of species extinction. Conservation Biology, 29, 452-462. |
| [6] |
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biological Conservation, 61, 1-10.
DOI URL |
| [7] |
Forest F, Grenyer R, Rouget M, Davies TJ, Cowling RM, Faith DP, Balmford A, Manning JC, Proches S, Bank M, Reeves G, Hedderson TAJ, Savolainen V (2007) Preserving the evolutionary potential of floras in biodiversity hotspots. Nature, 445, 757-760.
DOI URL |
| [8] | Gao LM, Liu J, Cai J, Yang JB, Zhang T, Li DZ (2012) A synopsis of technical notes on the standards for plant DNA barcoding. Plant Diversity and Resources, 34, 592-606. (in Chinese with English abstract) |
| [高连明, 刘杰, 蔡杰, 杨俊波, 张挺, 李德铢 (2012) 关于植物DNA条形码研究技术规范. 植物分类与资源学报, 34, 592-606.] | |
| [9] | Ge XJ (2015) Application of DNA barcoding in phylofloristics study. Biodiversity Science, 23, 295-296. (in Chinese) |
| [葛学军 (2015) DNA条形码在植物系统发育区系学研究中的应用. 生物多样性, 23, 295-296.] | |
| [10] |
Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM (2007) Mammals on the EDGE: Conservation priorities based on threat and phylogeny. PLoS ONE, 2, e296.
DOI URL |
| [11] | IUCN (2017) The IUCN Red List of Threatened Species. Version 2017-3. . (accessed on 2017 -12-05 |
| [12] |
Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26, 1463-1464.
DOI URL |
| [13] |
Klein C, Wilson K, Watts M, Stein J, Berry S, Carwardine J, Smith MS, Mackey B, Possingham H (2009) Incorporating ecological and evolutionary processes into continental-scale conservation planning. Ecological Applications, 19, 206-217.
DOI URL |
| [14] |
Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Berminghan E (2009) Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Sciences, USA, 106, 18621-18626.
DOI URL |
| [15] |
Laity T, Laffan SW, González-Orozco CE, Faith DP, Rosauer DF, Byrne M, Miller JT, Crayn D, Costion C, Moritz CC, Newport K (2015) Phylodiversity to inform conservation policy: An Australian example. Science of The Total Environment, 534, 131-143.
DOI URL PMID |
| [16] |
Larkin MA, Blackshields G, Brown NP, Chenna R, McGetti-gan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947-2948.
DOI URL |
| [17] |
Li HM, Aide TM, Ma YX, Liu WJ, Cao M (2007) Demand for rubber is causing the loss of high diversity rain forest in SW China. Plant Conservation and Biodiversity, 16, 1731-1745.
DOI URL |
| [18] | Li HW, Li J, Huang PH, Wei FN, Cui HB, van der Werff H (2008) Lauraceae. In: Flora of China, Vol. 7 (eds Wu ZY, Raven PH, Hong DY), pp. 102-254. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis. |
| [19] |
Liu ZF, Ci XQ, Li L, Li HW, Conran JG, Li J (2017) DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China. PLoS ONE, 12, e0175788.
DOI URL PMID |
| [20] |
Lyubetsky V, Piel WH, Quandt D (2014) Current advances in molecular phylogenetics. BioMed Research International, 2014, 596746.
DOI URL PMID |
| [21] |
Mace GM, Norris K, Fitter AH (2012) Biodiversity and ecosystem services: A multilayered relationship. Trends in Ecology & Evolution, 27, 19-26.
DOI URL PMID |
| [22] | Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop, 14, 1-8. |
| [23] |
Mishler BD, Knerr N, González-Orozco CE, Thornhill AH, Laffan SW, Miller JT (2014) Phylogenetic measures of biodiversity and neo- and paleo-endemism in Australian Acacia. Nature Communications, 5, 4473.
DOI URL PMID |
| [24] |
Mooers A?, Atkins RA (2003) Indonesia’s threatened birds: Over 500 million years of evolutionary heritage at risk. Animal Conservation, 6, 183-188.
DOI URL |
| [25] |
Moritz C (2002) Strategies to protect biological diversity and the evolutionary processes that sustain it. Systematic Biology, 51, 238-254.
DOI URL |
| [26] |
Oliver TH, Heard MS, Isaac NJB, Roy DB, Procter D, Eigenbrod F, Freckleton R, Hector A, Orme CD, Petchey OL, Proen?a V, Raffaelli D, Suttle KB, Mace GM, Martín-López B, Woodcock BA, Bullock JM (2015) Biodiversity and resilience of ecosystem functions. Trends in Ecology & Evolution, 30, 673-684.
DOI URL PMID |
| [27] |
Pollock LJ, Rosauer DF, Thornhill AH, Kujala H, Crisp MD, Miller JT, McCarthy MA (2015) Phylogenetic diversity meets conservation policy: Small areas are key to preserving eucalypt lineages. Philosophical Transactions of the Royal Society B: Biological Sciences, 370, 20140007.
DOI URL PMID |
| [28] |
Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter, 15, 8-15.
DOI URL |
| [29] |
Qian H, Jin Y, Ricklefs RE (2017) Phylogenetic diversity anomaly in angiosperms between eastern Asia and eastern North America. Proceedings of the National Academy of Sciences, USA, 114, 11452-11457.
DOI URL PMID |
| [30] |
Redding DW, Mooers A? (2006) Incorporating evolutionary measures into conservation prioritization. Conservation Biology, 20, 1670-1678.
DOI URL PMID |
| [31] | R Development Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.. (accessed on 2017-11-30 |
| [32] | Rodrigues ASL, Brooks TM, GastonKJ (2005) Integrating phylogenetic diversity in the selection of priority areas for conservation: Does it make a difference? In: Phylogeny and Conservation (eds Purvis A, Gittleman JL, Brooks T), pp. 101-119. Cambridge University Press, London. |
| [33] |
Rodrigues ASL, Gaston KJ (2002) Maximising phylogenetic diversity in the selection of networks of conservation areas. Biological Conservation, 105, 103-111.
DOI URL |
| [34] |
Rolland J, Cadotte MW, Davies J, Devictor V, Lavergne S, Mouquet N, Pavoine S, Rodrigues A, Thuiller W, Turcati L, Winter M, Zupan L, Jabot F, Morlon H (2012) Using phylogenies in conservation: New perspectives. Biology Letters, 8, 692-694.
DOI URL PMID |
| [35] |
Soutullo A, Dodsworth S, Heard SB, Mooers A? (2005) Distribution and correlates of carnivore phylogenetic diversity across the Americas. Animal Conservation, 8, 249-258.
DOI URL |
| [36] |
Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood- based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688-2690.
DOI URL PMID |
| [37] | Wang S, Xie Y (2004) China Species Red List, Vol.1: Red List. Higher Education Press, Beijing. (in Chinese) |
| [汪松, 解焱 (2004) 中国物种红色名录, 第1卷: 红色名录. 高等教育出版社, 北京. ] | |
| [38] |
Webb CO, Ackerly DD, Kembel SW (2008) Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics, 24, 2098-2100.
DOI URL PMID |
| [39] |
Winter M, Devictor V, Schweiger O (2013) Phylogenetic diversity and nature conservation: Where are we? Trends in Ecology & Evolution, 28, 199-204.
DOI URL PMID |
| [40] | Xishuangbanna Nature Reserve Comprehensive Investigation Group(1987) Xishuangbanna National Nature Reserve Comprehensive Investigation Reports. Yunnan Science and Technology Press, Kunming. (in Chinese) |
| [西双版纳自然保护区综合考察团(1987) 西双版纳自然保护区综合考察报告集. 云南科技出版社, 昆明.] | |
| [41] | Xishuangbanna National Nature Reserve Management Bureau, Yunnan Institute of Forest Inventory and Planning(2005) Xishuangbanna National Nature Reserve. Yunnan Education Publishing House, Kunming. (in Chinese) |
| [西双版纳国家级自然保护区管理局, 云南省林业调查规划院(2005) 西双版纳国家级自然保护区. 云南教育出版社, 昆明.] | |
| [42] |
Yek SH, Willliams SE, Burwell CJ, Robson SKA, Crozier RH (2009) Ground dwelling ants as surrogates for establishing conservation priorities in the Australian wet tropics. Journal of Insect Science, 9, 12.
DOI URL PMID |
| [43] | Zhu H, Wang H, Li BG, Zhou SS, Zhang JH (2015) Studies on the forest vegetation of Xishuangbanna. Plant Science Journal, 33, 641-726. (in Chinese with English abstract) |
| [朱华, 王洪, 李保贵, 周仕顺, 张建侯 (2015) 西双版纳森林植被研究. 植物科学学报, 33, 641-726.] | |
| [44] | Zhu H, Yan LC (2012) Native seed plants in Xishuangbanna of Yunnan. Science Press, Beijing. (in Chinese) |
| [朱华, 闫丽春 (2012) 云南西双版纳野生种子植物. 科学出版社, 北京.] |
| [1] | Jingjing Zhang, Wenbin Huang, Yiting Chen, Zepeng Yang, Weiye Ke, Zhaojie Peng, Shichao Wei, Zhiwei Zhang, Yisi Hu, Wenhua Yu, Wenliang Zhou. Reef-building coral diversity and distribution characteristics in the National Nature Reserve for Marine Ecology of Guangdong Nanpeng Islands [J]. Biodiv Sci, 2025, 33(4): 24424-. |
| [2] | Guo Yutong, Li Sucui, Wang Zhi, Xie Yan, Yang Xue, Zhou Guangjin, You Chunhe, Zhu Saning, Gao Jixi. Coverage and distribution of national key protected wild species in China’s nature reserves [J]. Biodiv Sci, 2025, 33(3): 24423-. |
| [3] | Rui Qu, Zhenjun Zuo, Youxin Wang, Liangjian Zhang, Zhigang Wu, Xiujuan Qiao, Zhong Wang. The biogeochemical niche based on elementome and its applications in different ecosystems [J]. Biodiv Sci, 2024, 32(4): 23378-. |
| [4] | Xuemeng Li, Jibao Jiang, Zenglu Zhang, Xiaojing Liu, Yali Wang, Yizhao Wu, Yinsheng Li, Jiangping Qiu, Qi Zhao. Earthworm biodiversity and its influencing factors in Baotianman National Nature Reserve [J]. Biodiv Sci, 2024, 32(4): 23352-. |
| [5] | Qifan Wang, Xiaohui Liu, Ziwei Zhu, Lei Liu, Xinxue Wang, Xuyang Ji, Shaochun Zhou, Zidong Zhang, Hongyu Dong, Minghai Zhang. Mammal and avian diversity in Beijicun National Nature Reserve, Heilongjiang Province, China [J]. Biodiv Sci, 2024, 32(4): 24024-. |
| [6] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [7] | Di Suo, Ruoxi Yu, Yuanhui Li, Jiliang Xu. Problem review and optimization path of local legislation in nature reserves in China based on empirical analysis [J]. Biodiv Sci, 2024, 32(2): 23287-. |
| [8] | Xiaolin Liu, Yougui Wu, Minhua Zhang, Xiaorong Chen, Zhicheng Zhu, Dingyun Chen, Shu Dong, Buhang Li, Bingyang Ding, Yu Liu. Community composition and structure of a 25-ha forest dynamics plot of subtropical forest in Baishanzu, Zhejiang Province [J]. Biodiv Sci, 2024, 32(2): 23294-. |
| [9] | Xiaolong Huang, Bingshun Meng, Haibo Li, Wei Ran, Wei Yang, Cheng Wang, Bo Xie, Xu Zhang, Jingcheng Ran, Mingming Zhang. Interspecific associations between Rhinopithecus brelichi and its sympatric species using infrared cameras [J]. Biodiv Sci, 2024, 32(2): 23402-. |
| [10] | Xianglin Yang, Caiyun Zhao, Junsheng Li, Fangfang Chong, Wenjin Li. Invasive plant species lead to a more clustered community phylogenetic structure: An analysis of herbaceous plants in Guangxi’s national nature reserves [J]. Biodiv Sci, 2024, 32(11): 24175-. |
| [11] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [12] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [13] | Ruirui Mao, Tuo Shen, Hui Li, Linchu Tian, Hairong Tan, Lirong Lu, Xiaogang Wu, Zongji Fan, Guoyi Wu, Jie Li, Yong Wu, Bicheng Zhu, Zhishu Xiao. A dataset of call characteristics of anuran from the Chebaling National Nature Reserve, Guangdong Province [J]. Biodiv Sci, 2024, 32(10): 24356-. |
| [14] | Guofa Cui. Discussion and suggestions on several key issues in the integration and optimization of protected areas [J]. Biodiv Sci, 2023, 31(9): 22447-. |
| [15] | Chao Xing, Yi Lin, Zhiqiang Zhou, Lianjun Zhao, Shiwei Jiang, Zhenzhen Lin, Jiliang Xu, Xiangjiang Zhan. The establishment of terrestrial vertebrate genetic resource bank and species identification based on DNA barcoding in Wanglang National Nature Reserve [J]. Biodiv Sci, 2023, 31(7): 22661-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()