



Biodiv Sci ›› 2016, Vol. 24 ›› Issue (6): 617-628. DOI: 10.17520/biods.2016054 cstr: 32101.14.biods.2016054
• Original Papers: Plant Diversity • Previous Articles Next Articles
Gexi Xu1, Zuomin Shi1,2,*( ), Jingchao Tang1, Han Xu3, Huai Yang4, Shirong Liu1, Yide Li3, Mingxian Lin4
), Jingchao Tang1, Han Xu3, Huai Yang4, Shirong Liu1, Yide Li3, Mingxian Lin4
Received:2016-02-25
Accepted:2016-06-05
Online:2016-06-20
Published:2016-06-20
Contact:
Shi Zuomin
Gexi Xu, Zuomin Shi, Jingchao Tang, Han Xu, Huai Yang, Shirong Liu, Yide Li, Mingxian Lin. Effects of species abundance and size classes on assessing community phylogenetic structure: a case study in Jianfengling tropical montane rainforest[J]. Biodiv Sci, 2016, 24(6): 617-628.
| 径级 Size classes | 植株数量 Individuals | 种数 Species | 种百分比 Species percentage (%) | 属数 Genus | 属百分比 Genus percentage (%) | 科数 Families | 科百分比 Family pergentage (%) |
|---|---|---|---|---|---|---|---|
| DBH≥15 cm | 1,689 (7.48%) | 146 | 61.86 | 82 | 64.06 | 42 | 75.00 |
| 5 cm≤DBH<15 cm | 3,302 (14.64%) | 191 | 80.93 | 109 | 85.16 | 54 | 96.43 |
| 1 cm≤DBH<5 cm | 17,570 (77.88%) | 226 | 95.76 | 123 | 96.09 | 54 | 96.43 |
| DBH≥1 cm | 22,561 | 236 | 100 | 128 | 100 | 56 | 100 |
Table 1 Distribution of species, genus and families of trees and shrubs in different DBH size classes in Jianfengling tropical montane rainforest community
| 径级 Size classes | 植株数量 Individuals | 种数 Species | 种百分比 Species percentage (%) | 属数 Genus | 属百分比 Genus percentage (%) | 科数 Families | 科百分比 Family pergentage (%) |
|---|---|---|---|---|---|---|---|
| DBH≥15 cm | 1,689 (7.48%) | 146 | 61.86 | 82 | 64.06 | 42 | 75.00 |
| 5 cm≤DBH<15 cm | 3,302 (14.64%) | 191 | 80.93 | 109 | 85.16 | 54 | 96.43 |
| 1 cm≤DBH<5 cm | 17,570 (77.88%) | 226 | 95.76 | 123 | 96.09 | 54 | 96.43 |
| DBH≥1 cm | 22,561 | 236 | 100 | 128 | 100 | 56 | 100 |
| 系统发育多样性指数 Phylogenetic diversity indices | 径级 Size classes | |||
|---|---|---|---|---|
| DBH≥15 cm | 5 cm≤DBH<15 cm | 1 cm≤DBH<5 cm | DBH≥1 cm | |
| MPD vs. AW-MPD | 0.797*** | 0.805*** | 0.473*** | 0.465*** |
| MNTD vs. AW-MNTD | 0.895*** | 0.888*** | 0.725*** | 0.828*** |
| NRI vs. AW-NRI | 0.876*** | 0.893*** | 0.621*** | 0.597*** |
| NTI vs. AW-NTI | 0.149 | 0.406*** | 0.542*** | 0.687*** |
Table 2 Pearson correlation of phylogenetic diversity indices with and without species weighted abundance
| 系统发育多样性指数 Phylogenetic diversity indices | 径级 Size classes | |||
|---|---|---|---|---|
| DBH≥15 cm | 5 cm≤DBH<15 cm | 1 cm≤DBH<5 cm | DBH≥1 cm | |
| MPD vs. AW-MPD | 0.797*** | 0.805*** | 0.473*** | 0.465*** |
| MNTD vs. AW-MNTD | 0.895*** | 0.888*** | 0.725*** | 0.828*** |
| NRI vs. AW-NRI | 0.876*** | 0.893*** | 0.621*** | 0.597*** |
| NTI vs. AW-NTI | 0.149 | 0.406*** | 0.542*** | 0.687*** |
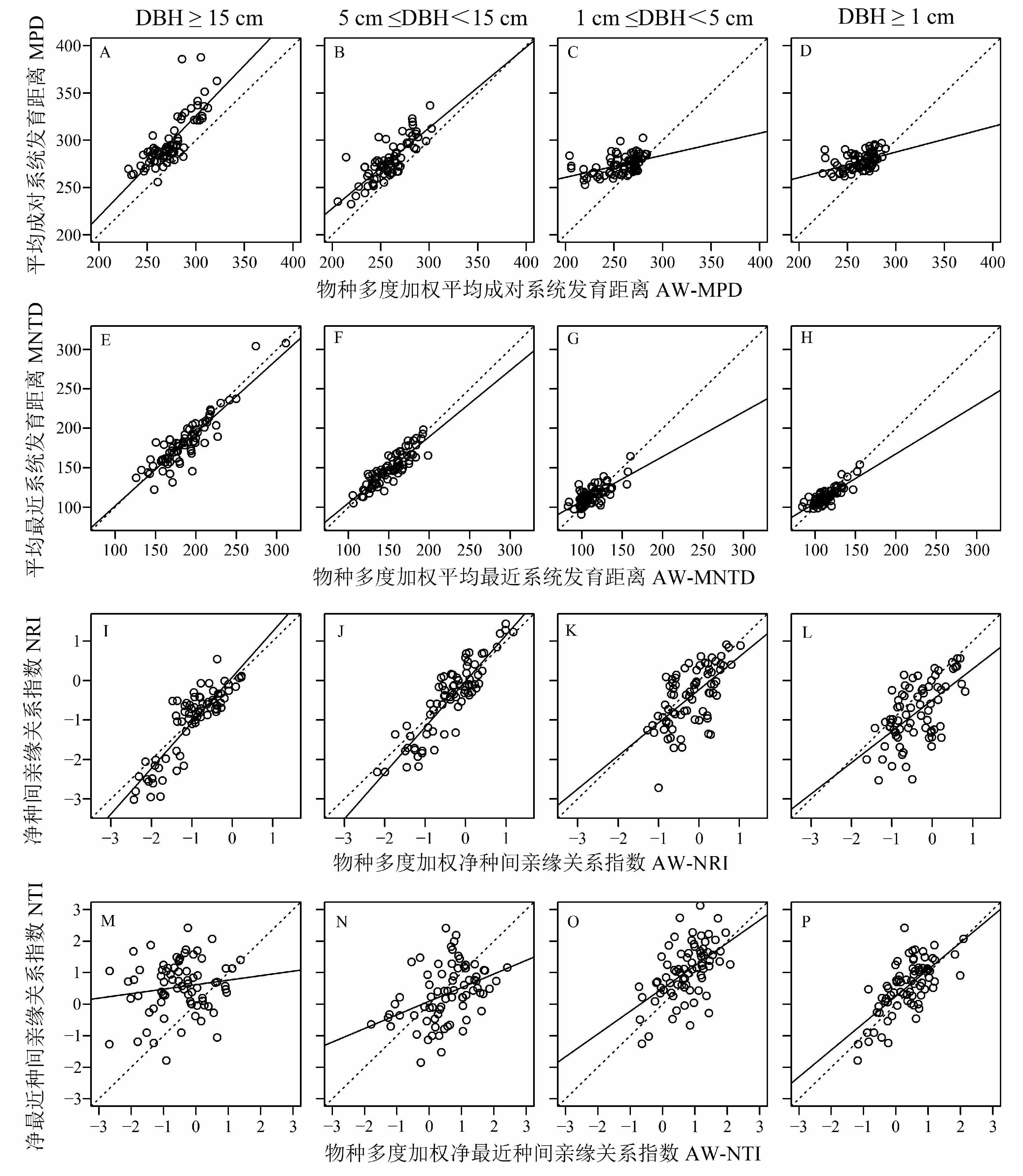
Fig. 1 Effects of species weighted abundance on phylogenetic diversity indices. The dash lines are 1:1 lines, and the solid lines are simulated lines based on simple linear regression. Scatter points distributed on the left side of the dash lines indicate overestimation of phylogenetic diversity, while those on the right side indicate underestimation of phylogenetic diversity. MPD, Mean pairwise distance; AW-MPD, Abundance weighted MPD; MNTD, Mean nearest taxon distance; AW-MNTD, Abundance weighted MNTD; NRI, Net relatedness index; AW-NRI, Abundance weighted NRI; NTI, Nearest taxon index; AW-NTI, Abundance weighted NTI.
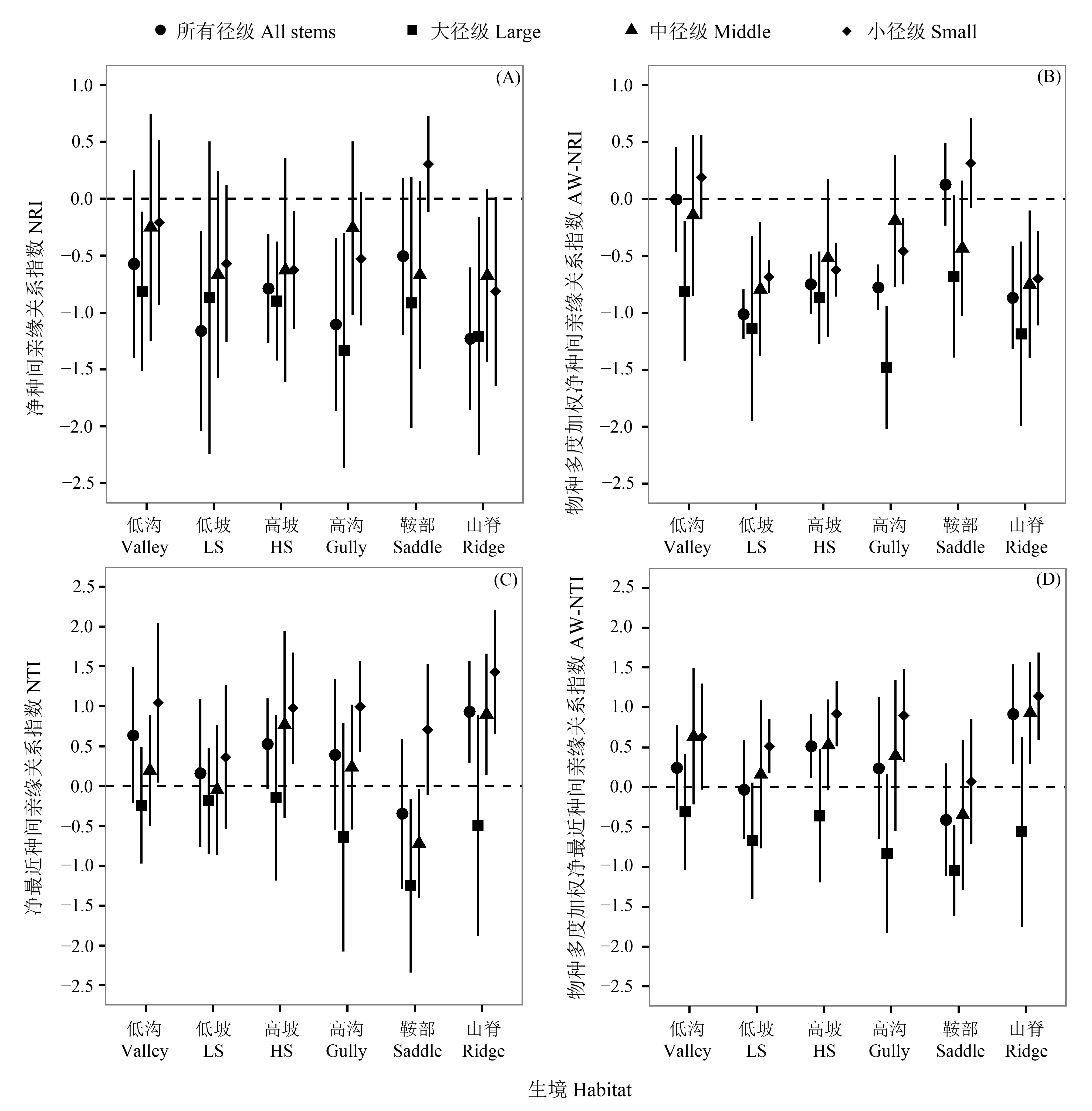
Fig. 2 The patterns (Mean±SD) of phylogenetic dispersion (NRI and NTI) with and without species weighted abundance for woody plants at different size classes in six habitat types at the spatial scale of 20 m×20 m. LS, Low slope; HS, High slope. Size classes: Small, 1 cm≤DBH<5 cm; Middle, 5 cm≤DBH<15 cm; Large, DBH≥15 cm.
| 生境 Habitat | 径级 Size classes | ||||
|---|---|---|---|---|---|
| DBH≥15 cm | 5 cm≤DBH<15 cm | 1 cm≤DBH<5 cm | DBH≥1 cm | ||
| 净种间亲缘关系指数 Net relatedness index (NRI) | 低沟 Valley | -0.81±0.70Ab | -0.25±1.00Aa | -0.21±0.73ABa | -0.57±0.82A |
| 低坡 Low slope | -0.87±1.37Aa | -0.66±0.91Aa | -0.57±0.69ABa | -1.16±0.88A | |
| 高坡 High slope | -0.90±0.52Aa | -0.63±0.98Aa | -0.62±0.52ABa | -0.79±0.48A | |
| 高沟 Gully | -1.33±1.03Ab | -0.26±0.76Aab | -0.53±0.59ABa | -1.10±0.76A | |
| 鞍部 Saddle | -0.91±1.10Aa | -0.67±0.91Aa | 0.30±0.42Aa | -0.51±0.69A | |
| 山脊 Ridge | -1.21±1.05Aa | -0.68±0.76Aa | -0.81±0.83Ba | -1.23±0.63A | |
| 物种多度加权净种间亲缘关系指数 Abundance weighted net relatedness index (AW-NRI) | 低沟 Valley | -0.81±0.61Ab | -0.14±0.71Aa | 0.19±0.37Aa | -0.01±0.46A |
| 低坡 Low slope | -1.13±0.81Aa | -0.79±0.58Aa | -0.68±0.71Ba | -1.01±0.22B | |
| 高坡 High slope | -0.87±0.40Aa | -0.52±0.69Aa | -0.62±0.24Ba | -0.75±0.26B | |
| 高沟 Gully | -1.48±0.54Ab | -0.19±0.58Aa | -0.46±0.29Ba | -0.78±0.20B | |
| 鞍部 Saddle | -0.68±0.71Ab | -0.43±0.59Aa | 0.31±0.40Aa | -0.13±0.36A | |
| 山脊 Ridge | -1.19±0.81Aa | -0.46±0.29Aa | -0.87±0.45Ba | -0.78±0.20B | |
| 净最近种间亲缘关系指数 Nearest taxon index (NTI) | 低沟 Valley | -0.24±0.73Ac | 0.20±0.69ABbc | 1.04±1.00Aa | 0.64±0.85AB |
| 低坡 Low slope | -0.18±0.66Aa | -0.04±0.82ABa | 0.36±0.90Aa | 0.16±0.93AB | |
| 高坡 High slope | -0.15±1.04Ab | 0.77±1.17Aa | 0.98±0.70Aa | 0.53±0.57AB | |
| 高沟 Gully | -0.64±1.44Ab | 0.24±0.78ABa | 1.00±0.57Aa | 0.39±0.95AB | |
| 鞍部 Saddle | -1.25±1.09Ab | -0.72±0.68Ba | 0.71±0.82Aa | -0.35±0.94B | |
| 山脊 Ridge | -0.49±1.38Ab | 0.90±0.76Aa | 1.43±0.78Aa | 0.93±0.64A | |
| 物种多度加权净最近种间亲缘关系指数 Abundance weighted nearest taxon index (AW-NTI) | 低沟 Valley | -0.31±0.73Ab | 0.64±0.85ABa | 0.63±0.66ABa | 0.25±0.53BC |
| 低坡 Low slope | -0.67±0.73Aa | 0.16±0.93ABa | 0.52±0.34ABa | -0.03±0.62BC | |
| 高坡 High slope | -0.36±0.84Ab | 0.53±0.57ABa | 0.92±0.41ABa | 0.51±0.40AB | |
| 高沟 Gully | -0.83±1.00Ab | 0.39±0.95ABa | 0.90±0.58ABa | 0.24±0.89BC | |
| 鞍部 Saddle | -1.04±0.57Aa | -0.35±0.94Ba | 0.07±0.79Ba | -0.41±0.71C | |
| 山脊 Ridge | -0.56±1.19Ab | 0.93±0.64Aa | 1.14±0.55Aa | 0.92±0.62A | |
Table 3 Tukey multi-comparison for NRI and NTI (mean±SD) with and without species weighted abundance among different DBH size classes and habitat types.
| 生境 Habitat | 径级 Size classes | ||||
|---|---|---|---|---|---|
| DBH≥15 cm | 5 cm≤DBH<15 cm | 1 cm≤DBH<5 cm | DBH≥1 cm | ||
| 净种间亲缘关系指数 Net relatedness index (NRI) | 低沟 Valley | -0.81±0.70Ab | -0.25±1.00Aa | -0.21±0.73ABa | -0.57±0.82A |
| 低坡 Low slope | -0.87±1.37Aa | -0.66±0.91Aa | -0.57±0.69ABa | -1.16±0.88A | |
| 高坡 High slope | -0.90±0.52Aa | -0.63±0.98Aa | -0.62±0.52ABa | -0.79±0.48A | |
| 高沟 Gully | -1.33±1.03Ab | -0.26±0.76Aab | -0.53±0.59ABa | -1.10±0.76A | |
| 鞍部 Saddle | -0.91±1.10Aa | -0.67±0.91Aa | 0.30±0.42Aa | -0.51±0.69A | |
| 山脊 Ridge | -1.21±1.05Aa | -0.68±0.76Aa | -0.81±0.83Ba | -1.23±0.63A | |
| 物种多度加权净种间亲缘关系指数 Abundance weighted net relatedness index (AW-NRI) | 低沟 Valley | -0.81±0.61Ab | -0.14±0.71Aa | 0.19±0.37Aa | -0.01±0.46A |
| 低坡 Low slope | -1.13±0.81Aa | -0.79±0.58Aa | -0.68±0.71Ba | -1.01±0.22B | |
| 高坡 High slope | -0.87±0.40Aa | -0.52±0.69Aa | -0.62±0.24Ba | -0.75±0.26B | |
| 高沟 Gully | -1.48±0.54Ab | -0.19±0.58Aa | -0.46±0.29Ba | -0.78±0.20B | |
| 鞍部 Saddle | -0.68±0.71Ab | -0.43±0.59Aa | 0.31±0.40Aa | -0.13±0.36A | |
| 山脊 Ridge | -1.19±0.81Aa | -0.46±0.29Aa | -0.87±0.45Ba | -0.78±0.20B | |
| 净最近种间亲缘关系指数 Nearest taxon index (NTI) | 低沟 Valley | -0.24±0.73Ac | 0.20±0.69ABbc | 1.04±1.00Aa | 0.64±0.85AB |
| 低坡 Low slope | -0.18±0.66Aa | -0.04±0.82ABa | 0.36±0.90Aa | 0.16±0.93AB | |
| 高坡 High slope | -0.15±1.04Ab | 0.77±1.17Aa | 0.98±0.70Aa | 0.53±0.57AB | |
| 高沟 Gully | -0.64±1.44Ab | 0.24±0.78ABa | 1.00±0.57Aa | 0.39±0.95AB | |
| 鞍部 Saddle | -1.25±1.09Ab | -0.72±0.68Ba | 0.71±0.82Aa | -0.35±0.94B | |
| 山脊 Ridge | -0.49±1.38Ab | 0.90±0.76Aa | 1.43±0.78Aa | 0.93±0.64A | |
| 物种多度加权净最近种间亲缘关系指数 Abundance weighted nearest taxon index (AW-NTI) | 低沟 Valley | -0.31±0.73Ab | 0.64±0.85ABa | 0.63±0.66ABa | 0.25±0.53BC |
| 低坡 Low slope | -0.67±0.73Aa | 0.16±0.93ABa | 0.52±0.34ABa | -0.03±0.62BC | |
| 高坡 High slope | -0.36±0.84Ab | 0.53±0.57ABa | 0.92±0.41ABa | 0.51±0.40AB | |
| 高沟 Gully | -0.83±1.00Ab | 0.39±0.95ABa | 0.90±0.58ABa | 0.24±0.89BC | |
| 鞍部 Saddle | -1.04±0.57Aa | -0.35±0.94Ba | 0.07±0.79Ba | -0.41±0.71C | |
| 山脊 Ridge | -0.56±1.19Ab | 0.93±0.64Aa | 1.14±0.55Aa | 0.92±0.62A | |
| [12] | Erickson DL, Jones FA, Swenson NG, Pei NC, Bourg NA, Chen WN, Davies SJ, Ge XJ, Hao ZQ, Howe RW, Huang CL, Larson AJ, Lum SKY, Lutz JA, Ma KP, Meegaskumbura M, Mi XC, Parker JD, Sun IF, Wright SJ, Wolf AT, Ye WH, Xing DL, Zimmerman JK, Kress WJ (2014) Comparative evolutionary diversity and phylogenetic structure across multiple forest dynamics plots: a mega-phylogeny approach. Frontiers in Genetics, 5, 358. |
| [13] | Guo ZG, Liu HX, Sun XG, Cheng GD (2003) Characteristics of species diversity of plant communities in the upper reaches of Bailong River. Acta Phytoecologica Sinica, 27, 388-395. (in Chinese with English abstract) |
| [郭正刚, 刘慧霞, 孙学刚, 程国栋 (2003) 白龙江上游地区森林植物群落物种多样性的研究. 植物生态学报, 27, 388-395.] | |
| [14] | He JS, Chen WL, Jiang MX, Jin YX, Hu D, Lu P (1998) Plant species diversity of the degraded ecosystems in the Three Gorges region. Acta Ecologica Sinica, 18, 399-407. (in Chinese with English abstract) |
| [贺金生, 陈伟烈, 江明喜, 金兴义, 胡东, 路鹏 (1998) 长江三峡地区退化生态系统植物群落物种多样性特征. 生态学报, 18, 399-407.] | |
| [15] | Helmus MR, Bland TJ, Williams CK, Ives AR (2007) Phylogenetic measures of biodiversity. The American Naturalist, 169, E68-E83. |
| [16] | Hubbell SP (2001) The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32). Princeton University Press, New Jersey. |
| [17] | Huston M (1979) A general hypothesis of species diversity. The American Naturalist, 113, 81-101. |
| [18] | Jiang YX, Lu JP (1991) Forest Ecosystem of Tropical Forest of Jianfengling Mountain, Hainan Island, China. Science Press, Beijing. (in Chinese) |
| [蒋有绪, 卢俊培 (1991) 中国海南岛尖峰岭热带林生态系统. 科学出版社, 北京.] | |
| [19] | Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26, 1463-1464. |
| [20] | Kreft H, Jetz W (2007) Global patterns and determinants of vascular plant diversity. Proceedings of the National Academy of Sciences, USA, 104, 5925-5930. |
| [21] | Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E (2009) Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Sciences, USA, 106, 18621-18626. |
| [22] | Lasky JR, Uriarte M, Boukili VK, Chazdon RL (2014) Trait-mediated assembly processes predict successional changes in community diversity of tropical forests. Proceedings of the National Academy of Sciences, USA, 111, 5616-5621. |
| [23] | Letcher SG, Lasky JR, Chazdon RL, Norden N, Wright SJ, Meave JA, Pérez-García EA, Muñoz R, Romero-Pérez E, Andrade A, Andrade JL, Balvanera P, Becknell JM, Bentos TV, Bhaskar R, Bongers F, Boukili V, Brancalion PHS, César RG, Clark DA, Clark DB, Craven D, DeFrancesco A, Dupuy JM, Finegan B, González-Jiménez E, Hall JS, Harms KE, Hernández-Stefanoni JL, Hietz P, Kennard D, Killeen TJ, Laurance SG, Lebrija-Trejos EE, Lohbeck M, Martínez-Ramos M, Massoca PES, Mesquita RCG, Mora F, Muscarella R, Paz H, Pineda-García F, Powers JS, Quesada-Monge R, Rodrigues RR, Sandor ME, Sanaphre-Villanueva L, Schüller E, Swenson NG, Tauro A, Uriarte M, van Breugel M, Vargas-Ramírez O, Viani RAG, Wendt AL, Williamson GB (2015) Environmental gradients and the evolution of successional habitat specialization: a test case with 14 Neotropical forest sites. Journal of Ecology, 103, 1276-1290. |
| [24] | Li YD, Chen BF, Zhou GY, Wu ZM, Zeng QB, Luo TS, Huang SN, Xie MD, Huang Q (2002) Research and Conservation of Tropical Forest and the Biodiversity: A Species Reference to Hainan Island, China. China Forestry Publishing House, Beijing. (in Chinese) |
| [李意德, 陈步峰, 周光益, 吴仲民, 曾庆波, 骆土寿, 黄世能, 谢明东, 黄全 (2002)中国海南岛热带森林及其生物多样性保护研究. 中国林业出版社, 北京.] | |
| [25] | Li YD, Xu H, Luo TS, Chen DX, Lin MX (2012) Permant Monitoring and Research Dataset of Chinese Ecosystem: Forest Ecosystem: Jianfengling Station (Bio-Species Checklist). China Agriculture Press, Beijing. (in Chinese) |
| [李意德, 许涵, 骆土寿, 陈德祥, 林明献 (2012) 中国生态系统定位观测与研究数据集: 森林生态系统卷: 海南尖峰岭站(生物物种数据集). 中国农业出版社, 北京] | |
| [26] | Losos JB (1994) An approach to the analysis of comparative data when a phylogeny is unavailable or incomplete. Systematic Biology, 43, 117-123. |
| [27] | Losos JB (2008) Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecology Letters, 11, 995-1003. |
| [28] | Ma KP (2013) Studies on biodiversity and ecosystem function via manipulation experiments. Biodiversity Science, 21, 247-248. (in Chinese) |
| [马克平 (2013) 生物多样性与生态系统功能的实验研究. 生物多样性, 21, 247-248.] | |
| [29] | Magallón S, Castillo A (2009) Angiosperm diversification through time. American Journal of Botany, 96, 349-365. |
| [30] | Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289-290. |
| [31] | Pool TK, Grenouillet G, Villéger S (2014) Species contribute differently to the taxonomic, functional, and phylogenetic alpha and beta diversity of freshwater fish communities. Diversity and Distributions, 20, 1235-1244. |
| [32] | R Core Team (2015) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Stastical Computing.2014. . |
| [33] | Rao CR (1982) Diversity and dissimilarity coefficients: a unified approach. Theoretical Population Biology, 21, 24-43. |
| [34] | Swenson NG (2011) The role of evolutionary processes in producing biodiversity patterns, and the interrelationships between taxonomic, functional and phylogenetic biodiversity. American Journal of Botany, 98, 472-480. |
| [1] | Andersen KM, Turner BL, Dalling JW (2014) Seedling performance trade-offs influencing habitat filtering along a soil nutrient gradient in a tropical forest. Ecology, 95, 3399-3413. |
| [2] | Anderson T, Lachance M, Starmer W (2004) The relationship of phylogeny to community structure: the cactus yeast community. The American Naturalist, 164, 709-721. |
| [35] | Swenson NG (2013) The assembly of tropical tree communities —the advances and shortcomings of phylogenetic and functional trait analyses. Ecography, 36, 264-276. |
| [36] | Swenson NG (2014) Functional and Phylogenetic Ecology in R. Springer Science & Business Media, New York. |
| [3] | Anderson-Teixeira KJ, Davies SJ, Bennett AC, Gonzalez-Akre EB, Muller-Landau HC, Joseph Wright S, Salim AK, Zambrano AMA, Alonso A, Baltzer JL, Basset Y, Bourg NA, Broadbent EN, Brockelman WY, Bunyavejchewin S, Burslem DFRP, Butt N, Cao M, Cardenas D, Chuyong GB, Clay K, Cordell S, Dattaraja HS, Deng XB, Detto M, Du XJ, Duque A, Erikson DL, Ewango CEN, Fischer GA, Fletcher C, Foster RB, Giardina CP, Gilbert GS, Gunatilleke N, Gunatilleke S, Hao ZQ, Hargrove WW, Hart TB, Hau BCH, He FL, Hoffman FM, Howe RW, Hubbell SP, Inman-Narahari FM, Jansen PA, Jiang MX, Johnson DJ, Kanzaki M, Kassim AR, Kenfack D, Kibet S, Kinnaird MF, Korte L, Kral K, Kumar J, Larson AJ, Li YD, Li XK, Liu SR, Lum SKY, Lutz JA, Ma KP, Maddalena DM, Makana J-R, Malhi Y, Marthews T, Serudin RM, McMahon SM, McShea WJ, Memiaghe HR, Mi XC, Mizuno T, Morecroft M, Myers JA, Novotny V, de Oliveira AA, Ong PS, Orwig DA, Ostertag R, Ouden JD, Parker GG, Phillips RP, Sack L, Sainge MN, Sang WG, Sri-ngernyuang K, Sukumar R, Sun IF, Sungpalee W, Suresh HS, Tan S, Thomas SC, Thomas DW, Thompson J, Turner BL, Uriarte M, Valencia R, Vallejo MI, Vicentini A, Vrška T, Wang XH, Wang XG, Weiblen G, Wolf A, Xu H, Yap S, Zimmerman J (2014) CTFS-ForestGEO: a worldwide network monitoring forests in an era of global change. Global Change Biology, 21, 528-549. |
| [4] | Bremer B, Bremer K, Chase M, Fay M, Reveal J, Soltis D, Soltis P, Stevens P (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society, 161, 105-121. |
| [5] | Castro SA, Escobedo VM, Aranda J, Carvallo GO (2014) Evaluating Darwin’s naturalization hypothesis in experimental plant assemblages: phylogenetic relationships do not determine colonization success. PLoS ONE, 9, e105535. |
| [6] | CBOL Plant Working Group (2009) A DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA, 106, 12794-12797. |
| [37] | Swenson NG, Enquist BJ, Thompson J, Zimmerman JK (2007) The influence of spatial and size scale on phylogenetic relatedness in tropical forest communities. Ecology, 88, 1770-1780. |
| [38] | Swenson NG, Erickson DL, Mi XC, Bourg NA, Forero- Montaña J, Ge XJ, Howe R, Lake JK, Liu XJ, Ma KP, Pei NC, Thompson J, Uriarte M, Wolf A, Wright SJ, Ye WH, Zhang JL, Zimmerman JK, Kress WJ (2012) Phylogenetic and functional alpha and beta diversity in temperate and tropical tree communities. Ecology, 93, S112-S125. |
| [39] | Tilman D, Knops J, Wedin D, Reich P, Ritchie M, Siemann E (1997) The influence of functional diversity and composition on ecosystem processes. Science, 277, 1300-1302. |
| [7] | Chamberlain S, Vázquez DP, Carvalheiro L, Elle E, Vamosi JC (2014) Phylogenetic tree shape and the structure of mutualistic networks. Journal of Ecology, 102, 1234-1243. |
| [8] | Condit R (1998) Tropical Forest Census Plots: Methods and Results from Barro Colorado Island, Panama and a Comparison With Other Plots. Springer Science & Business Media, New York. |
| [40] | Vellend M, Cornwell WK, Magnuson-Ford K, Mooers AØ (2011) Measuring phylogenetic biodiversity. In: Biological Diversity: Frontiers in Measurement and Assessment (eds Magurran A, McGill B), pp. 194-207. Oxford University Press, Oxford, UK. |
| [41] | Webb CO (2000) Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. The American Naturalist, 156, 143-155. |
| [9] | Cooper N, Jetz W, Freckleton RP (2010) Phylogenetic comparative approaches for studying niche conservatism. Journal of Evolutionary Biology, 23, 2529-2539. |
| [10] | Donoghue MJ (2008) A phylogenetic perspective on the distribution of plant diversity. Proceedings of the National Academy of Sciences, USA, 105, 11549-11555. |
| [42] | Webb CO, Ackerly DD, Kembel SW (2008) Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics, 24, 2098-2100. |
| [43] | Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Annual Review of Ecology and Systematics, 33, 475-505. |
| [44] | Webb CO, Donoghue MJ (2005) Phylomatic: tree assembly for applied phylogenetics. Molecular Ecology Notes, 5, 181-183. |
| [45] | Webb CO, Losos JB, Agrawal AA (2006) Integrating phylogenies into community ecology. Ecology, 87, S1-S2. |
| [46] | Wickham H (2009) Ggplot2: Elegent Graphics for Data Analysis. Springer Science & Business Media, New York. |
| [47] | Xu H, Li YD, Lin MX, Wu JH, Luo TS, Zhou Z, Chen DX, Yang H, Li GJ, Liu SR (2015) Community characteristics of a 60 ha dynamics plot in the tropical montane rain forest in Jianfengling, Hainan Island. Biodiversity Science, 23, 192-201. (in Chinese with English abstract) |
| [许涵, 李意德, 林明献, 吴建辉, 骆土寿, 周璋, 陈德祥, 杨怀, 李广建, 刘世荣 (2015) 海南尖峰岭热带山地雨林60 ha动态监测样地群落结构特征. 生物多样性, 23, 192-201.] | |
| [48] | Yang J, Ci XQ, Lu MN, Zhang GC, Cao M, Li J, Lin LX (2014) Functional traits of tree species with phylogenetic signal co-vary with environmental niches in two large forest dynamics plots. Journal of Plant Ecology, 7, 115-125. |
| [49] | Yang J, Zhang GC, Ci XQ, Swenson NG, Cao M, Sha LQ, Li J, Baskin CC, Slik JWF, Lin LX (2013) Functional and phylogenetic assembly in a Chinese tropical tree community across size classes, spatial scales and habitats. Functional Ecology, 28, 520-529. |
| [50] | Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, Royer DL, Soltis DE, Stevens PF, Westoby M, Wright IJ, Aarssen L, Bertin RI, Calaminus A, Govaerts R, Hemmings F, Leishman MR, Oleksyn J, Soltis PS, Swenson NG, Warman L, Beaulieu JM (2014) Three keys to the radiation of angiosperms into freezing environments. Nature, 506, 89-92. |
| [51] | Zeng QB, Li YD, Chen BF, Wu ZM, Zhou GY (1997) Research and Management of Tropical Forest Ecosystem. China Forestry Publishing House, Beijing. (in Chinese) |
| [曾庆波, 李意德, 陈步峰, 吴仲民, 周光益 (1997) 热带森林生态系统研究与管理. 中国林业出版社, 北京.] | |
| [52] | Zhu Y, Comita LS, Hubbell SP, Ma KP (2015) Conspecific and phylogenetic density-dependent survival differs across life stages in a tropical forest. Journal of Ecology, 103, 957-966. |
| [11] | Eastman JM, Harmon LJ, Tank DC (2013) Congruification: support for time scaling large phylogenetic trees. Methods in Ecology and Evolution, 4, 688-691. |
| [1] | Li Yanpeng, Chen Jie, Lu Chunyang, Xu Han. Community characteristics of a 64-ha secondary forest dynamics plot in a tropical montane rainforest in Jianfengling, Hainan [J]. Biodiv Sci, 2025, 33(2): 24445-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [4] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [5] | Shuhan Yang, He Wang, Lei Chen, Yingfei Liao, Guang Yan, Yining Wu, Hongfei Zou. Effects of heterogeneous habitat on soil nematode community characteristics in the Songnen Plain [J]. Biodiv Sci, 2024, 32(1): 23295-. |
| [6] | Minghui Wang, Zhaoquan Chen, Shuaifeng Li, Xiaobo Huang, Xuedong Lang, Zihan Hu, Ruiguang Shang, Wande Liu. Spatial pattern of dominant species with different seed dispersal modes in a monsoon evergreen broad-leaved forest in Pu’er, Yunnan Province [J]. Biodiv Sci, 2023, 31(9): 23147-. |
| [7] | Yanqiu Xie, Hui Huang, Chunxiao Wang, Yaqin He, Yixuan Jiang, Zilin Liu, Chuanyuan Deng, Yushan Zheng. Determinants of species-area relationship and species richness of coastal endemic plants in the Fujian islands [J]. Biodiv Sci, 2023, 31(5): 22345-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Wenbo Yan, Yanni Mo, Zhigao Zeng, Shaoliang Xue, Qi Wang, Chunsheng Liang, Zhuli Huang, Wen Luo, Daye Liu, Shiqin Mo, Xiaoguang Li, Lu Liang, Kunpeng Du. Distribution and conservation status of Chinese pangolin (Manis pentadactyla) in Jianfengling, Hainan [J]. Biodiv Sci, 2022, 30(6): 22106-. |
| [10] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [11] | Bo Chen, Lan Jiang, Ziyang Xie, Yangdi Li, Jiaxuan Li, Mengjia Li, Chensi Wei, Cong Xing, Jinfu Liu, Zhongsheng He. Taxonomic and phylogenetic diversity of plants in a Castanopsis kawakamiinatural forest [J]. Biodiv Sci, 2021, 29(4): 439-448. |
| [12] | Guijun Yang, Min Wang, Yichun Yang, Xinyun Li, Xinpu Wang. Distribution patterns and environmental interpretation of beetle species richness in Helan Mountain of northern China [J]. Biodiv Sci, 2019, 27(12): 1309-1319. |
| [13] | Dexin Sun, Xiang Liu, Shurong Zhou. Dynamical changes of diversity and community assembly during recovery from a plant functional group removal experiment in the alpine meadow [J]. Biodiv Sci, 2018, 26(7): 655-666. |
| [14] | Yu Zhang, Gang Feng. Distribution pattern and mechanism of insect species diversity in Inner Mongolia [J]. Biodiv Sci, 2018, 26(7): 701-706. |
| [15] | Hou Qinxi, Ci Xiuqin, Liu Zhifang, Xu Wumei, Li Jie. Assessment of the evolutionary history of Lauraceae in Xishuangbanna National Nature Reserve using DNA barcoding [J]. Biodiv Sci, 2018, 26(3): 217-228. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()