



生物多样性 ›› 2010, Vol. 18 ›› Issue (2): 129-136. DOI: 10.3724/SP.J.1003.2010.134 cstr: 32101.14.SP.J.1003.2010.134
收稿日期:2009-07-07
接受日期:2009-11-16
出版日期:2010-03-20
发布日期:2010-03-20
通讯作者:
张道远
基金资助:
Yan Liu1,2, Daoyuan Zhang1,*( )
)
Received:2009-07-07
Accepted:2009-11-16
Online:2010-03-20
Published:2010-03-20
Contact:
Daoyuan Zhang
摘要:
采用空间自相关分析方法对准噶尔无叶豆(Eremosparton songoricum)5个居群遗传变异的空间结构进行了研究。根据8对ISSR引物扩增的多态性位点, 选择了基因频率在30-70%的ISSR位点, 运用GenAlEx 6和SAAP4.3软件以及地理距离间隔方法分别计算了5个居群的空间自相关系数。结果表明: 准噶尔无叶豆居群具有明显的空间遗传结构, 绝大多数ISSR位点变异呈非随机分布。位于沙漠腹地的B、D居群和位于沙漠东缘的F居群的遗传变异呈现渐变模式, 分别在7 m, 7 m及9 m范围内个体呈现显著正相关, 推测与克隆繁殖占绝对优势、有性更新罕见发生有关, 而克隆大小的差别则主要由微生境差异所引起。位于沙漠北缘的G居群的遗传变异呈现双向渐变模式, 在3 m内个体呈现显著正相关, 这与该居群实生幼苗增补密切相关, 但花粉和种子的有限散布所导致的距离隔离对该居群空间遗传结构产生了重要影响。而沙漠腹地的A居群呈现衰退模式, 是因为小居群增加了近交的几率, 并引起了遗传漂变。
刘燕, 张道远, 杨红兰 (2010) 准噶尔无叶豆5个自然居群ISSR遗传变异的空间自相关分析. 生物多样性, 18, 129-136. DOI: 10.3724/SP.J.1003.2010.134.
Yan Liu, Daoyuan Zhang (2010) Spatial genetic structure in five natural populations of Eremosparton songoricum as revealed by ISSR analysis. Biodiversity Science, 18, 129-136. DOI: 10.3724/SP.J.1003.2010.134.
| 居群 Population | 经纬度 Location | 海拔(m) Altitude | 群丛类型 Associatioin type | 生境 Habitat |
|---|---|---|---|---|
| A | 44°38'N 88°16'E | 548 | 准噶尔无叶豆 + 蛇麻黄群丛 Eremosparton songoricum+ Ephedra distachyaassociation | 南北向沙垄下部风蚀流沙地(位于沙漠腹地) Wind erosion sand surface of the lower south-north dune (located in hinterland desert, small population) |
| B | 44°46'N 88°16'E | 605 | 准噶尔无叶豆 + 沙蒿 + 三芒草群丛 Eremosparton songoricum + Artemisia arenaria + Aristida pennataassociation | 垄间低地及沙丘中下部(位于沙漠腹地) Interdune and middle-lower part of the dune (located in hinterland desert) |
| D | 44°58'N 88°25'E | 647 | 准噶尔无叶豆群丛 Eremosparton songoricumassociation | 道路和引水工程等建设区周围或两侧的缓起伏沙垄(沙丘)上(位于沙漠腹地) On dunes beside the road or encircle the transferring water project (located in hinterland desert) |
| F | 44°14'N 90°04'E | 718 | 准噶尔无叶豆 + 三芒草 + 一年生长营养期植物群系丛 Eremosparton songoricum + Aristida pennata + annual plant association | 垄底部至缓起伏的垄顶(位于沙漠东缘) From the bottom to the top of dune (located in east of desert) |
| G | 46°21'N 88°38'E | 794 | 准噶尔无叶豆 + 沙蒿群丛 Eremosparton songoricum + Artemisia arenaria association | 平缓流沙地(位于沙漠北缘, 紧邻乌仑谷湖) Flat wind erosion sand dune (located in north of desert, beside Wulungu Lake) |
表1 古尔班通古特沙漠准噶尔无叶豆5个居群的分布地点、群丛类型及生境特点
Table 1 Distribution, association type and habitats of five Eremosparton songoricum populations in Gurbantunggut Desert
| 居群 Population | 经纬度 Location | 海拔(m) Altitude | 群丛类型 Associatioin type | 生境 Habitat |
|---|---|---|---|---|
| A | 44°38'N 88°16'E | 548 | 准噶尔无叶豆 + 蛇麻黄群丛 Eremosparton songoricum+ Ephedra distachyaassociation | 南北向沙垄下部风蚀流沙地(位于沙漠腹地) Wind erosion sand surface of the lower south-north dune (located in hinterland desert, small population) |
| B | 44°46'N 88°16'E | 605 | 准噶尔无叶豆 + 沙蒿 + 三芒草群丛 Eremosparton songoricum + Artemisia arenaria + Aristida pennataassociation | 垄间低地及沙丘中下部(位于沙漠腹地) Interdune and middle-lower part of the dune (located in hinterland desert) |
| D | 44°58'N 88°25'E | 647 | 准噶尔无叶豆群丛 Eremosparton songoricumassociation | 道路和引水工程等建设区周围或两侧的缓起伏沙垄(沙丘)上(位于沙漠腹地) On dunes beside the road or encircle the transferring water project (located in hinterland desert) |
| F | 44°14'N 90°04'E | 718 | 准噶尔无叶豆 + 三芒草 + 一年生长营养期植物群系丛 Eremosparton songoricum + Aristida pennata + annual plant association | 垄底部至缓起伏的垄顶(位于沙漠东缘) From the bottom to the top of dune (located in east of desert) |
| G | 46°21'N 88°38'E | 794 | 准噶尔无叶豆 + 沙蒿群丛 Eremosparton songoricum + Artemisia arenaria association | 平缓流沙地(位于沙漠北缘, 紧邻乌仑谷湖) Flat wind erosion sand dune (located in north of desert, beside Wulungu Lake) |
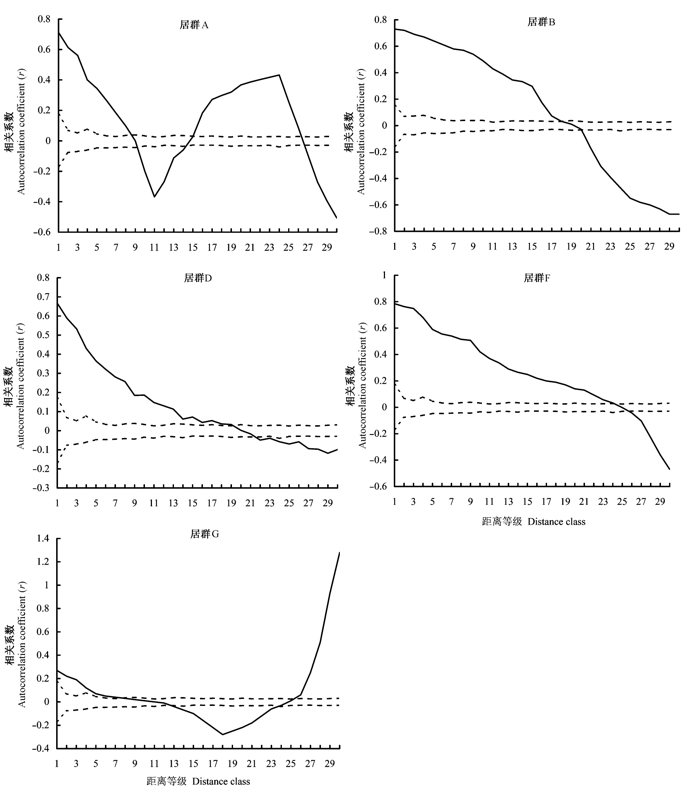
图1 准噶尔无叶豆5个居群遗传变异的空间自相关图。依据ISSR多态位点频率变异在30个距离等级上的空间自相关系数(r)平均值得出的自相关图。实线代表自相关系数, 虚线代表95%的置信区间。
Fig. 1 Correlogram using mean value of Moran’sIof ISSR loci frequency variance for five Eremosparton songoricum populations. Solid lines represent the correlation coefficient (r) and dashed lines represent the 95% confidence intervals.
| 距离等级 Distance class | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1.0 | 2.0 | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 | 8.0 | 9.0 | 10.0 | |
| 居群A Population A | ||||||||||
| 正相关(%) | 31(93.9) | 19(57.6) | 4(12.1) | 6(18.2) | 11(33.3) | 19(57.6) | 19(57.6) | 24(72.7) | 8(24.2) | 2(6.1) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 13(39.4) | 17(51.5) | 18(54.5) | 16(48.5) | 11(33.3) | 8(24.2) | 7(21.2) | 21(63.6) | 26(78.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 93.9 | 97.0 | 63.6 | 72.7 | 81.8 | 90.9 | 81.8 | 93.9 | 87.9 | 84.8 |
| Significant correlation (%) | ||||||||||
| 居群B Population B | ||||||||||
| 正相关(%) | 25(100) | 19(76.0) | 17(68.0) | 15(60.0) | 16(64.0) | 12(48.0) | 8(32.0) | 3(12.0) | 2(8.0) | 1(4.0) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 1(4.0) | 4(16.0) | 7(28.0) | 8(32.0) | 10(40.0) | 17(68.0) | 20(80.0) | 23(92.0) | 21(84.0) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 80.0 | 84.0 | 88.0 | 96.0 | 88.0 | 100.0 | 92.0 | 100.0 | 88.0 |
| Significant correlation (%) | ||||||||||
| 居群D Population D | ||||||||||
| 正相关(%) | 27(100) | 23(85.2) | 16(59.3) | 14(51.9) | 14(51.9) | 13(48.1) | 10(37.0) | 8(29.6) | 5(18.5) | 6(22.2) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 3(11.1) | 9(33.3) | 13(48.1) | 12(44.4) | 12(44.4) | 15(55.6) | 18(66.7) | 19(70.4) | 20(74.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 96.3 | 92.6 | 100.0 | 96.3 | 92.6 | 92.6 | 96.3 | 88.9 | 96.3 |
| Significant correlation (%) | ||||||||||
| 居群F Population F | ||||||||||
| 正相关(%) | 28(100) | 19(67.9) | 20(71.4) | 15(53.6) | 17(60.7) | 11(39.3) | 15(53.6) | 14(50.0) | 10(35.7) | 4(14.3) |
| Positive correlation (%) | ||||||||||
| 负相关(比例) | 0(0) | 4(14.3) | 7(25.0) | 8(28.6) | 10(35.7) | 8(28.6) | 11(39.3) | 12(42.9) | 18(64.3) | 23(82.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 82.1 | 96.4 | 82.1 | 96.4 | 67.9 | 92.9 | 92.9 | 100.0 | 96.4 |
| Significant correlation (%) | ||||||||||
| 居群G Population G | ||||||||||
| 正相关(%) | 22(75.9) | 14(48.3) | 7(24.1) | 11(37.9) | 9(31.0) | 7(24.1) | 9(31.0) | 10(34.5) | 21(72.4) | 24(82.8) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 9(31.0) | 5(17.2) | 12(41.4) | 14(48.3) | 18(62.1) | 15(51.7) | 12(41.4) | 3(10.3) | 2(6.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 75.9 | 79.3 | 41.4 | 79.3 | 79.3 | 86.2 | 82.8 | 75.9 | 82.8 | 89.7 |
| Significant correlation (%) | ||||||||||
表2 准噶尔无叶豆5个居群内各距离等级中表现出显著相关的位点数(等距离间隔)及百分比
Table 2 Number of loci showing significant correlation in each distance class for Eremosparton songoricum in different populations (equal distance interval) and the percentage
| 距离等级 Distance class | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1.0 | 2.0 | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 | 8.0 | 9.0 | 10.0 | |
| 居群A Population A | ||||||||||
| 正相关(%) | 31(93.9) | 19(57.6) | 4(12.1) | 6(18.2) | 11(33.3) | 19(57.6) | 19(57.6) | 24(72.7) | 8(24.2) | 2(6.1) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 13(39.4) | 17(51.5) | 18(54.5) | 16(48.5) | 11(33.3) | 8(24.2) | 7(21.2) | 21(63.6) | 26(78.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 93.9 | 97.0 | 63.6 | 72.7 | 81.8 | 90.9 | 81.8 | 93.9 | 87.9 | 84.8 |
| Significant correlation (%) | ||||||||||
| 居群B Population B | ||||||||||
| 正相关(%) | 25(100) | 19(76.0) | 17(68.0) | 15(60.0) | 16(64.0) | 12(48.0) | 8(32.0) | 3(12.0) | 2(8.0) | 1(4.0) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 1(4.0) | 4(16.0) | 7(28.0) | 8(32.0) | 10(40.0) | 17(68.0) | 20(80.0) | 23(92.0) | 21(84.0) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 80.0 | 84.0 | 88.0 | 96.0 | 88.0 | 100.0 | 92.0 | 100.0 | 88.0 |
| Significant correlation (%) | ||||||||||
| 居群D Population D | ||||||||||
| 正相关(%) | 27(100) | 23(85.2) | 16(59.3) | 14(51.9) | 14(51.9) | 13(48.1) | 10(37.0) | 8(29.6) | 5(18.5) | 6(22.2) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 3(11.1) | 9(33.3) | 13(48.1) | 12(44.4) | 12(44.4) | 15(55.6) | 18(66.7) | 19(70.4) | 20(74.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 96.3 | 92.6 | 100.0 | 96.3 | 92.6 | 92.6 | 96.3 | 88.9 | 96.3 |
| Significant correlation (%) | ||||||||||
| 居群F Population F | ||||||||||
| 正相关(%) | 28(100) | 19(67.9) | 20(71.4) | 15(53.6) | 17(60.7) | 11(39.3) | 15(53.6) | 14(50.0) | 10(35.7) | 4(14.3) |
| Positive correlation (%) | ||||||||||
| 负相关(比例) | 0(0) | 4(14.3) | 7(25.0) | 8(28.6) | 10(35.7) | 8(28.6) | 11(39.3) | 12(42.9) | 18(64.3) | 23(82.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 82.1 | 96.4 | 82.1 | 96.4 | 67.9 | 92.9 | 92.9 | 100.0 | 96.4 |
| Significant correlation (%) | ||||||||||
| 居群G Population G | ||||||||||
| 正相关(%) | 22(75.9) | 14(48.3) | 7(24.1) | 11(37.9) | 9(31.0) | 7(24.1) | 9(31.0) | 10(34.5) | 21(72.4) | 24(82.8) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 9(31.0) | 5(17.2) | 12(41.4) | 14(48.3) | 18(62.1) | 15(51.7) | 12(41.4) | 3(10.3) | 2(6.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 75.9 | 79.3 | 41.4 | 79.3 | 79.3 | 86.2 | 82.8 | 75.9 | 82.8 | 89.7 |
| Significant correlation (%) | ||||||||||
| [1] | Васипьченко (1941) ФЛОРА СССР. Tomus Ⅺ (苏联植物志(第十一卷)), pp.310-311. (in Russian). |
| [2] |
Charlesworth B, Langley CH (1989) The population genetics of Drosophila transposable elements. Annual Review of Genetics, 23,251-287.
DOI URL PMID |
| [3] | Chen CD (陈昌笃), Zhang LY (张立运), Hu WK (胡文康) (1983) The basic characteristics of plant communities, flora and their distribution in the sandy district of Gurbantunggut. Acta Phytoecologica et Geobotanica Sinica (植物生态学与地植物学丛刊), 7,89-99. (in Chinese with English abstract) |
| [4] |
Chung MG, Chung JM, Chung MY, Epperson BK (2000) Spatial genetic distribution of allozyme polymorphisms following clonal and sexual reproduction in populations of Rhus javanica (Anacardiaceae) . Heredity, 84,178-185.
URL PMID |
| [5] |
Chung MY, Nason JD, Chung MG (2004) Spatial genetic structure in populations of the terrestrial orchid Cephalanthera longibracteata (Orchidaceae) . American Journal of Botany, 91,52-57.
DOI URL PMID |
| [6] |
Dong M (1995) Morphological responses to local light conditions of clonal herbs from contrasting habitats, and their modification due to physiological integration. Oecologia, 101,282-288.
DOI URL PMID |
| [7] | Eckert CG (2000) Contributions of autogamy and geitonogamy to self-fertilization in a mass-flowering, clonal plant. Ecology, 81,532-542. |
| [8] | Eckert CG, Barrett SCH (1993) Clonal reproduction and patterns of genotypic diversity in Decodon verticillatus (Lythraceae) . American Journal of Botany, 80,1175-1182. |
| [9] | Eckert CG (2002) The loss of sex in clonal plants. Evolutionary Ecology, 15,501-520. |
| [10] | Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics, 24,2l7-242. |
| [11] |
Epperson BK (1990) Spatial autocorrelation of genotypes under directional selection. Genetics, 124,757-771.
URL PMID |
| [12] |
Eriksson O (1993) Dynamics of genets in clonal plants. Trends in Ecology and Evolution, 8,313-316.
DOI URL PMID |
| [13] | Falk DA, Holsinger KE (1991) Genetics and Conservation of Rare Plants. Oxford University Press, Oxford, |
| [14] | Geburek T, Tripp-Knowles P (1994) Genetic architecture in buroak, Quercus macrocarpa (Fagaceae), inferred by means of spatial autocorrelation analysis, Plant Systematics and Evolution, 189,63-74. |
| [15] | Hamrick JL, Godt MJW, Murawski DA, Loveless MD (1991) Correlations between species traits and allozyme diversity:implications for conservation biology. In: In: Genetics and Conservation of Rare Plants (eds Falk DA, Holsinger KE)pp.75-86. Oxford University Press, New York. |
| [16] | Handel SN (1985) The intrusion of clonal growth patterns of plant breeding systems. The American Naturalist, 125,367-384. |
| [17] | Huenneke LF (1991) Ecological implications of genetic variation in plant populations. In: In: Genetics and Conservation of Rare Plants (eds Falk DA, Holsinger KE), pp.31-44. Oxford University Press, New York. |
| [18] |
Kelly BA, Hardy OJ, Bouvet JM (2004) Temporal and spatial genetic structure in Vitellaria paradoxa (shea tree) in an agroforestry system in southern Mali . Molecular Ecology, 13,1231-1240.
DOI URL PMID |
| [19] |
Lande R (1988) Genetics and demography in biological conservation. Science, 241,1455-1460.
DOI URL PMID |
| [20] | Leberg PL (1990) Genetic considerations in the design of introduction programs. Transactions of the North American Wildlife and Natural Resource Conference, 55,609-619. |
| [21] |
Levin DA, Kerster HW (1974) Gene flow in seed plants. Evolutionary Biology, 7,139-220.
DOI URL PMID |
| [22] | Li ZZ (李作洲), Lang P (郎萍), Huang HW (黄宏文) (2002) Spatial structure of allozyme frequencies in Castanea mollissima BI. Journal of Wuhan Botanical Research (武汉植物学研究), 20,165-170. (in Chinese with English abstract) |
| [23] | Li ZZ (李作洲), Gong JJ (龚俊杰), Wang Y (王瑛), Huang HW (黄宏文) (2003) Spatial structure of AFLP genetic diversity of remnant populations of Metasequoia glyptostroboides (Taxodiaceae) . Biodiversity Science (生物多样性), 11,265-275. (in Chinese with English abstract) |
| [24] | Liu YL (刘亚令), Li ZZ (李作洲), Zhang PF (张鹏飞), Jiang ZW (姜正旺), Huang HW (黄宏文) (2006) Spatial genetic structure in natural populations of two closely related Actinidia species (Actinidiaceae) as revealed by SSR analysis . Biodiversity Science (生物多样性), 14,421-434. (in Chinese with English abstract) |
| [25] | Loiselle BA, Sork VL, Nason J, Grnham C (1995) Spatial genetic structure of a tropical understory shrub. Psychotria offinalis (Rubiaceae). American Journal of Botany, 82,1420-1425. |
| [26] | Lu XY (陆雪莹), Zhang DY (张道远), Ma WB (马文宝) (2007) Genetic variation and clonal diversity in fragmented populations of the desert plant Eremosparton songoricum based on ISSR markers . Biodiversity Science (生物多样性), 15,282-291. (in Chinese with English abstract) |
| [27] | Ma WB (马文宝), Shi X (施翔), Zhang DY (张道远), Yin LK (尹林克) (2008) Flowering phenology and reproductive features of the rare plant Eremosparton songoricum in desert zone, Xinjiang, China . Journal of Plant Ecology (植物生态学报), 32,760-767. (in Chinese with English abstract) |
| [28] | Ma WB (马文宝), Zhang DY (张道远), Yin LK (尹林克) (2007) Variation of seed sizes of Eremosparton songoricum geographic populations and germination characteristics . Arid Land Geography (干旱区地理), 30,674-679. (in Chinese with English abstract) |
| [29] |
Marquardt PE, Epperson BK (2004) Spatial and population genetic structure of microsatellites in white pine. Molecular Ecology, 13,3305-33l5.
DOI URL PMID |
| [30] | Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6,288-295. |
| [31] | Shi X, Wang JC, Zhang DY, Gaskin J, Pan BR (2010a) Pollination ecology of the rare desert species Eremosparton songoricum (Fabaceae) . Australian Journal of Botany, 58,35-41. |
| [32] | Shi X, Wang JC, Zhang DY, Gaskin J, Pan BR (2010b) Pollen source and resource limitation to fruit production in the rare species Eremosparton songoricum (Fabaceae) . Nordic Journal of Botany, 28,1-7. |
| [33] |
Sokal RR, Jacquez GM, Wooten M (1989) Spatial autocorrelation analysis of migration and selection. Genetics, 121,845-855.
URL PMID |
| [34] | Sokal RR, Oden NL (1978) Spatial autocorrelation in biology. 1. Methodology. Biological Journal of the Linnean Society, 10,199-228. |
| [35] | Sutherland S, Delph LF (1984) On the importance of male fitness in plants: patterns of fruit-set. Ecology, 65,1093-1104. |
| [36] | Wang JC (王建成), Shi X (施翔), Zhang DY (张道远), Yin LK (尹林克) (2009) The morphological plasticity of Eremosparton songoricum along heterogeneous micro-habitats of continuous moisture gradient changes in sand duns . Acta Ecologica Sinica (生态学报), 29,3641-3648. (in Chinese with English abstract) |
| [37] | Warternberg D (1989) SAAP 4.3: A Spatial Autocorrelation Analysis Program.. Exeter Software, Setauket, New York. |
| [38] | Yin LK (尹林克), Tan LX (谭丽霞), Wang B (王兵) (2006) Rare and Endangered Higher Plants Endemic to Xinjiang (新疆珍稀濒危特有高等植物), pp.74-75. Xinjiang Scientific and Technical Publishing House, Urumq. (in Chinese) |
| [39] |
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution, 11,413-418.
DOI URL PMID |
| [40] | Zhang DY (张道远), Ma WB (马文宝), Shi X (施翔), Wang JC (王建成), Wang XY (王习勇) (2008) Distribution and bio-ecological characteristics of Eremosparton songoricum, a rare plant in Gurbantunggut desert . Journal of Desert Research (中国沙漠), 28,430-436. (in Chinese with English abstract) |
| [41] | Zhang DY (张道远), Wang JC (王建成), Shi X (施翔) (2009) Population quantitative dynamics of the rhizomatous woody clonal plant Eremosparton songoricum in China’s Gurbantunggut Desert . Chinese Journal of Plant Ecology (植物生态学报), 33,893-900. (in Chinese with English abstract) |
| [42] | Zhang LY (张立运), Hai Y (海鹰) (2002) Plant communities excluded in the book of “ The Vegetation and Its Utilization in Xinjiang”.Ⅰ. The desert plant communities . Arid Land Geography (干旱区地理), 25,84-89. (in Chinese with English abstract) |
| [1] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [2] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [3] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [4] | 向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧. 多基因联合揭示海南鲌的遗传结构与遗传多样性[J]. 生物多样性, 2021, 29(11): 1505-1512. |
| [5] | 张亚红, 贾会霞, 王志彬, 孙佩, 曹德美, 胡建军. 滇杨种群遗传多样性与遗传结构[J]. 生物多样性, 2019, 27(4): 355-365. |
| [6] | 张剑, 董路, 张雁云. 基于基因组SNPs的南极恩克斯堡岛阿德利企鹅繁殖种群的遗传结构[J]. 生物多样性, 2019, 27(12): 1291-1297. |
| [7] | 武星彤, 陈璐, 王敏求, 张原, 林雪莹, 李鑫玉, 周宏, 文亚峰. 丹霞梧桐群体遗传结构及其遗传分化[J]. 生物多样性, 2018, 26(11): 1168-1179. |
| [8] | 刘青青, 董志军. 基于线粒体COI基因分析钩手水母的群体遗传结构[J]. 生物多样性, 2018, 26(11): 1204-1211. |
| [9] | 梁健超, 丁志锋, 张春兰, 胡慧建, 朵海瑞, 唐虹. 青海三江源国家级自然保护区麦秀分区鸟类多样性空间格局及热点区域研究[J]. 生物多样性, 2017, 25(3): 294-303. |
| [10] | 熊敏, 田双, 张志荣, 范邓妹, 张志勇. 华木莲居群遗传结构与保护单元[J]. 生物多样性, 2014, 22(4): 476-484. |
| [11] | 杨爱红, 张金菊, 田华, 姚小洪, 黄宏文. 鹅掌楸贵州烂木山居群的微卫星遗传多样性及空间遗传结构[J]. 生物多样性, 2014, 22(3): 375-384. |
| [12] | 徐刚标, 梁艳, 蒋燚, 刘雄盛, 胡尚力, 肖玉菲, 郝博搏. 伯乐树种群遗传多样性及遗传结构[J]. 生物多样性, 2013, 21(6): 723-731. |
| [13] | 王德元, 彭婕, 陈雅静, 吕国胜, 张小平, 邵剑文. 毛茛叶报春的遗传多样性及遗传结构[J]. 生物多样性, 2013, 21(5): 601-609. |
| [14] | 李晶晶, 张杰, 胡自民, 段德麟. 加拿大-西北∇大西洋地区掌形藻的种群遗传结构与动态变化[J]. 生物多样性, 2013, 21(3): 306-314. |
| [15] | 阮咏梅, 张金菊, 姚小洪, 叶其刚. 黄梅秤锤树孤立居群的遗传多样性及其小尺度空间遗传结构[J]. 生物多样性, 2012, 20(4): 460-469. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn