



Biodiv Sci ›› 2023, Vol. 31 ›› Issue (11): 23170. DOI: 10.17520/biods.2023170 cstr: 32101.14.biods.2023170
• Original Papers: Microbial Diversity • Previous Articles Next Articles
Jie Tao1,2,3, Benqiang Li1,2,3, Jinghua Cheng1,2,3, Ying Shi1,2,3, Peihong Liu1, Guixia He4, Weijie Xu4, Huili Liu1,2,3,*( )
)
Received:2023-05-26
Accepted:2023-09-05
Online:2023-11-20
Published:2023-09-20
Contact:
* E-mail: About author:# Co-first authors
Jie Tao, Benqiang Li, Jinghua Cheng, Ying Shi, Peihong Liu, Guixia He, Weijie Xu, Huili Liu. Viral metagenome analysis of the viral community composition of the porcine diarrhea feaces[J]. Biodiv Sci, 2023, 31(11): 23170.
| 微生物名称 Microbial name | 百分比 Percentage (%) | ||
|---|---|---|---|
| 噬菌体 Phages | 31.55 | ||
| 未分类病毒 Unclassified viruses | 26.77 | ||
| DNA病毒 DNA viruses | 小环状DNA病毒科 Smacoviridae | 1.96 | 3.10 |
| 圆环病毒 Circoviridae | 0.63 | ||
| 类双生病毒科 Genomoviridae | 0.50 | ||
| 其他DNA病毒 Other DNA viruses | 0.01 | ||
| RNA病毒 RNA viruses | 星状病毒科 Astroviridae | 19.15 | 38.58 |
| 杯状病毒科 Caliciviridae | 15.15 | ||
| 小RNA病毒科 Picornaviridae | 2.78 | ||
| 小双节RNA病毒科 Picobirnaviridae | 1.35 | ||
| 呼肠孤病毒 Reoviridae | 0.07 | ||
| 其他RNA病毒 Other RNA viruses | 0.08 | ||
Table 1 Metagenomic analysis of porcine diarrhea feces
| 微生物名称 Microbial name | 百分比 Percentage (%) | ||
|---|---|---|---|
| 噬菌体 Phages | 31.55 | ||
| 未分类病毒 Unclassified viruses | 26.77 | ||
| DNA病毒 DNA viruses | 小环状DNA病毒科 Smacoviridae | 1.96 | 3.10 |
| 圆环病毒 Circoviridae | 0.63 | ||
| 类双生病毒科 Genomoviridae | 0.50 | ||
| 其他DNA病毒 Other DNA viruses | 0.01 | ||
| RNA病毒 RNA viruses | 星状病毒科 Astroviridae | 19.15 | 38.58 |
| 杯状病毒科 Caliciviridae | 15.15 | ||
| 小RNA病毒科 Picornaviridae | 2.78 | ||
| 小双节RNA病毒科 Picobirnaviridae | 1.35 | ||
| 呼肠孤病毒 Reoviridae | 0.07 | ||
| 其他RNA病毒 Other RNA viruses | 0.08 | ||
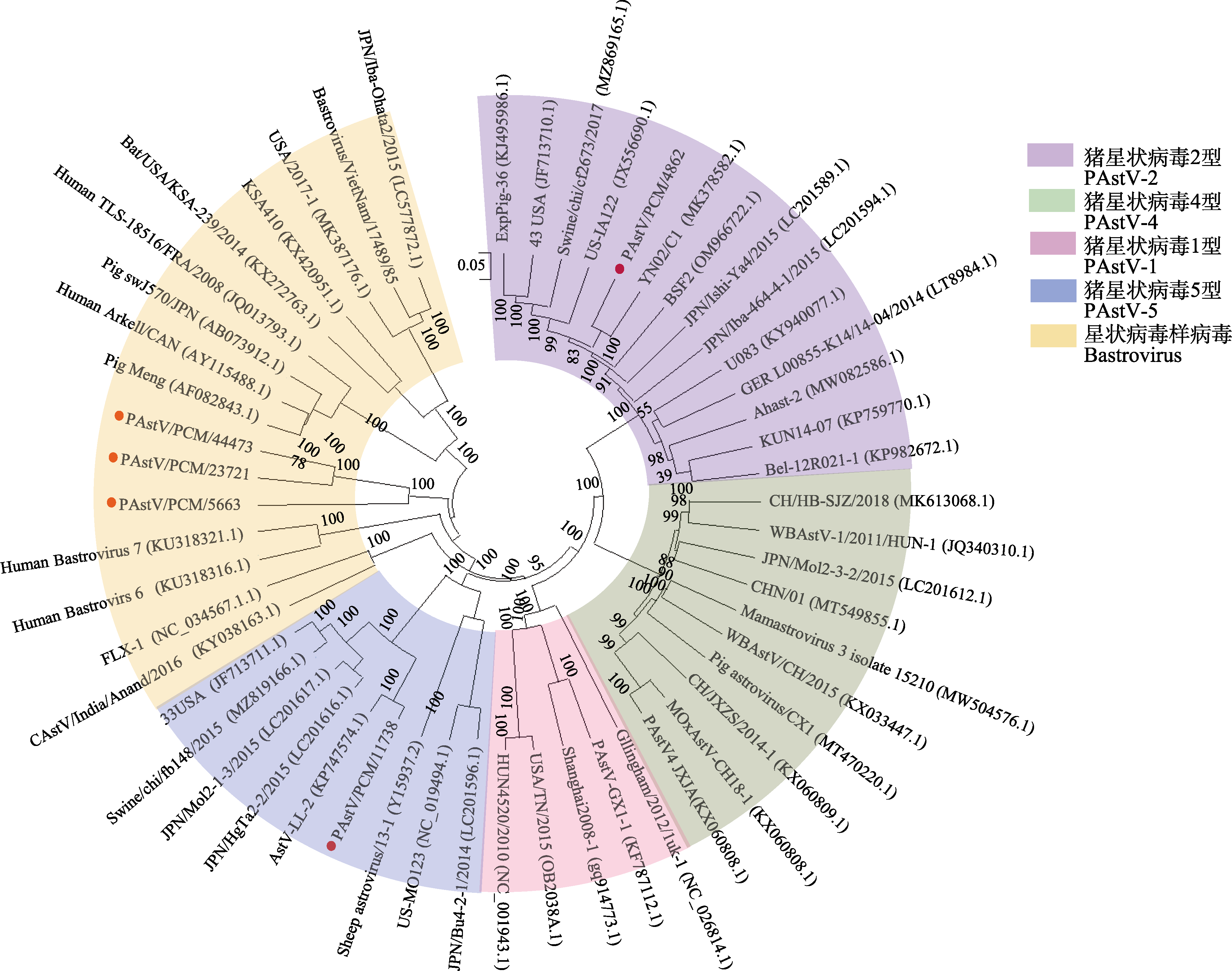
Fig. 2 Phylogenetic analysis of astrovirus based on the whole genome. Four PAstV genotypes (PAstV-1, PAstV-2, PAstV-3, PAstV-4 and PAstV-5) and bastrovirus were included in the phylogenetic tree. The red dots represent the viral sequences obtained in this study. 0.05 represents the scale of the phylogenetic tree; The values on the branches represent the Bootstrap value, indicating the confidence level.
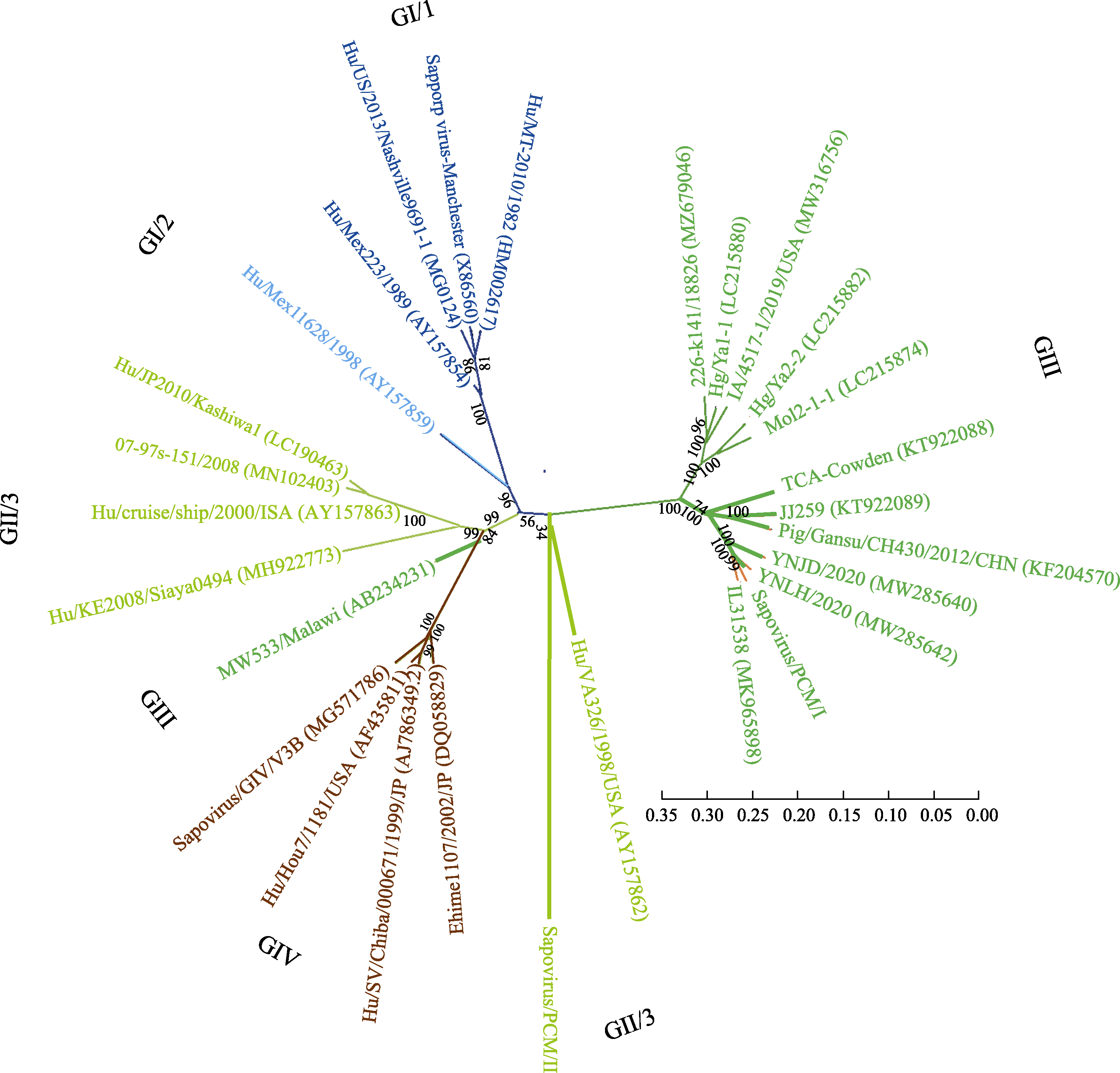
Fig. 3 Phylogenetic analysis of sapovirus based on the whole genome. Four genotypes of GI (GI/1 and GI/2), GII (GII/3), GIII and GIV were included in this phylogenetic tree. The ruler represents the scale of the phylogenetic tree; The values on the branches represent the confidence level.
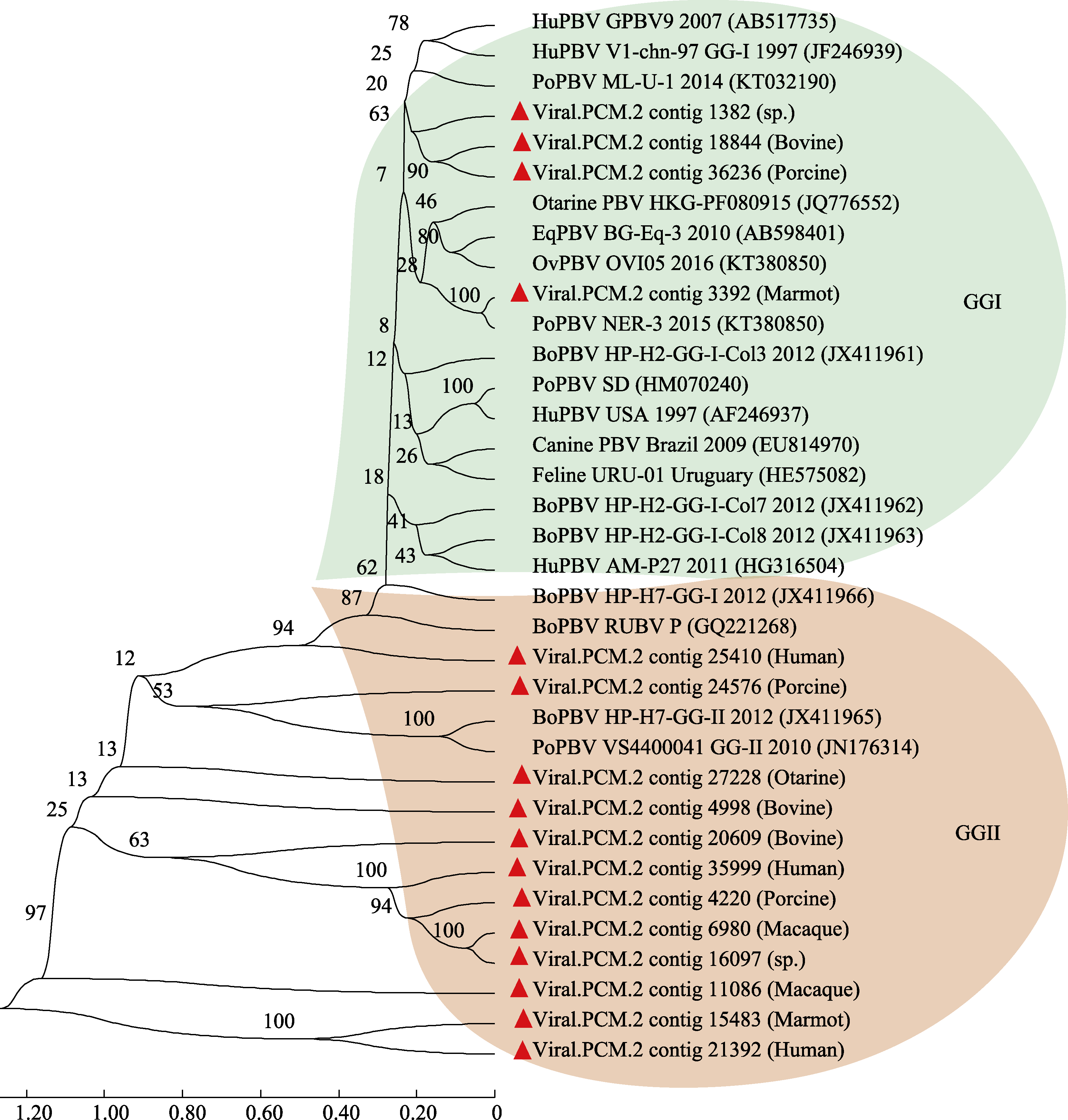
Fig. 4 Phylogenetic analysis of picobirnavirus based on the whole genome. Two genotypes of GGI and GGII were included in this phylogenetic tree. The ruler represents the scale of the phylogenetic tree; The values on the branches represent the confidence level.
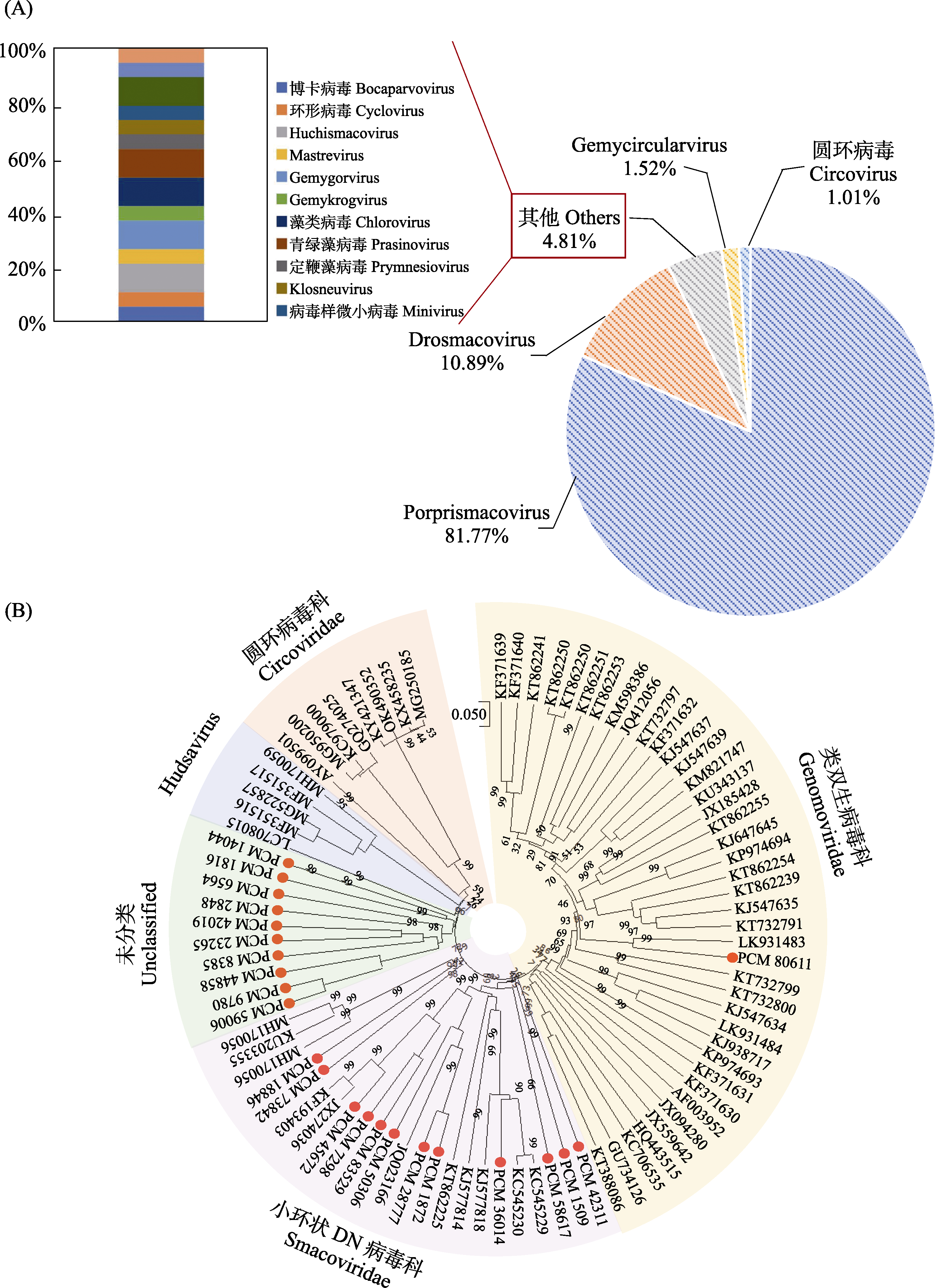
Fig. 5 Composition and phylogenetic analysis of DNA viruses. A, Composition analysis of DNA virus sequences; B, Phylogenetic analysis of CRESS-DNA viruses. 0.05 represents the distance scale (the scale of the phylogenetic tree); The values on the branches represent the bootstrap value.
| [1] |
Bai GH, Lin SC, Hsu YH, Chen SY (2022) The human virome: Viral metagenomics, relations with human diseases, and therapeutic applications. Viruses, 14, 278.
DOI URL |
| [2] |
Bauermann FV, Hause B, Buysse AR, Joshi LR, Diel DG (2019) Identification and genetic characterization of a porcine hepe-astrovirus (bastrovirus) in the United States. Archives of Virology, 164, 2321-2326.
DOI PMID |
| [3] |
Bhella D, Goodfellow IG (2011) The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor- induced flexibility and two distinct conformational changes in the capsid protein VP1. Journal of Virology, 85, 11381-11390.
DOI PMID |
| [4] |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114-2120.
DOI PMID |
| [5] |
Chen CY, Zhou YY, Fu H, Xiong XW, Fang SM, Jiang H, Wu JY, Yang H, Gao J, Huang LS (2021) Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nature Communications, 12, 1106.
DOI PMID |
| [6] |
Ganesh B, Bányai K, Martella V, Jakab F, Masachessi G, Kobayashi N (2012) Picobirnavirus infections: Viral persistence and zoonotic potential. Reviews in Medical Virology, 22, 245-256.
DOI PMID |
| [7] |
Guo JR, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, Pratama AA, Gazitúa MC, Vik D, Sullivan MB, Roux S (2021) VirSorter2: A multi-classifier, expert- guided approach to detect diverse DNA and RNA viruses. Microbiome, 9, 37.
DOI URL |
| [8] |
Hameed M, Liu K, Anwar MN, Wahaab A, Li CX, Di D, Wang X, Khan S, Xu JP, Li BB, Nawaz M, Shao DH, Qiu YF, Wei JC, Ma ZY (2020) A viral metagenomic analysis reveals rich viral abundance and diversity in mosquitoes from pig farms. Transboundary and Emerging Diseases, 67, 328-343.
DOI PMID |
| [9] |
Knox MA, Gedye KR, Hayman DS (2018) The challenges of analysing highly diverse Picobirnavirus sequence data. Viruses, 10, 685.
DOI URL |
| [10] |
Li DH, Luo RB, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW (2016) MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods, 102, 3-11.
DOI PMID |
| [11] |
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754-1760.
DOI PMID |
| [12] |
Liu TN, Liu CX, Liao JY, Xiong WJ, Xia JY, Xiao CT (2022) Identification and genomic characterization of a novel porcine CRESS DNA virus from a pig suffering from diarrhea in China. Archives of Virology, 167, 1355-1359.
DOI |
| [13] |
Liu X, Song CL, Liu YH, Qu KX, Bi JY, Bi JL, Wang YH, Yang Y, Sun JH, Guo ZG, Li GW, Liu JP, Yin GF (2022) High genetic diversity of porcine sapovirus from diarrheic piglets in Yunnan Province, China. Frontiers in Veterinary Science, 9, 854905.
DOI URL |
| [14] |
Martínez LC, Masachessi G, Carruyo G, Ferreyra LJ, Barril PA, Isa MB, Giordano MO, Ludert JE, Nates SV (2010) Picobirnavirus causes persistent infection in pigs. Infection, Genetics and Evolution, 10, 984-988.
DOI PMID |
| [15] |
Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC (2021) CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nature Biotechnology, 39, 578-585.
DOI PMID |
| [16] | Park YS, Kim S, Park DG, Kim DH, Yoon KW, Shin W, Han K (2019) Comparison of library construction kits for mRNA sequencing in the Illumina platform. Genes & Genomics, 41, 1233-1240. |
| [17] |
Pons JC, Paez-Espino D, Riera G, Ivanova N, Kyrpides NC, Llabrés M (2021) VPF-Class: Taxonomic assignment and host prediction of uncultivated viruses based on viral protein families. Bioinformatics, 37, 1805-1813.
DOI PMID |
| [18] |
Qiu M, Li SB, Xiao YZ, Li JX, Zhang YW, Li XS, Feng BH, Li C, Lin H, Zhu JZ, Chen NH (2022) Pathogenic and metagenomic evaluations reveal the correlations of porcine epidemic diarrhea virus, porcine kobuvirus and porcine astroviruses with neonatal piglet diarrhea. Microbial Pathogenesis, 170, 105703.
DOI URL |
| [19] | Shen XJ, Li JJ, Bian J, Zhao TT, Hua XG, Cui L (2019) Metagenomics analysis of feces from diarrhea piglets reveals the viral composition and epidemic characteristics of Coronavirus. Acta Veterinaria et Zootechnica Sinica, 50, 611-619. (in Chinese with English abstract) |
| [沈小娟, 李晶娇, 边疆, 赵婷婷, 华修国, 崔立 (2019) 腹泻仔猪肠道病毒组学分析. 畜牧兽医学报, 50, 611-619.] | |
| [20] |
Shi Y, Tao J, Li BQ, Shen XH, Cheng JH, Liu HL (2021) The gut viral metagenome analysis of domestic dogs captures snapshot of viral diversity and potential risk of coronavirus. Frontiers in Veterinary Science, 8, 695088.
DOI URL |
| [21] |
Shu X, Han F, Hu Y, Hao CL, Li ZY, Wei ZY, Zhang HL (2022) Co-infection of porcine deltacoronavirus and porcine epidemic diarrhoea virus alters gut microbiota diversity and composition in the colon of piglets. Virus Research, 322, 198954.
DOI URL |
| [22] |
Slavov SN (2022) Viral metagenomics for identification of emerging viruses in transfusion medicine. Viruses, 14, 2448.
DOI URL |
| [23] | Su MJ, Qi SS, Yang D, Guo DH, Yin BS, Sun DB (2020) Coinfection and genetic characterization of porcine astrovirus in diarrheic piglets in China from 2015 to 2018. Frontiers in Veterinary Science, 7, 462. |
| [24] | Villanova F, Milagres FAP, Brustulin R, Araújo ELL, Pandey RP, Raj VS, Deng XT, Delwart E, Luchs A, da Costa AC, Leal É (2022) A new circular single-stranded DNA virus related with howler monkey associated Porpris macovirus 1 detected in children with acute gastroenteritis. Viruses, 14, 1472. |
| [25] |
Wang LY, Marthaler D, Fredrickson R, Gauger PC, Zhang JQ, Burrough ER, Petznick T, Li GW (2020) Genetically divergent porcine sapovirus identified in pigs, United States. Transboundary and Emerging Diseases, 67, 18-28.
DOI PMID |
| [26] |
Wang QH, Han MG, Funk JA, Bowman G, Janies DA, Saif LJ (2005) Genetic diversity and recombination of porcine sapoviruses. Journal of Clinical Microbiology, 43, 5963-5972.
DOI URL |
| [27] |
Wang QH, Vlasova AN, Kenney SP, Saif LJ (2019) Emerging and re-emerging coronaviruses in pigs. Current Opinion in Virology, 34, 39-49.
DOI PMID |
| [28] | Wu Q, Zhang Y, Zheng HX, Yin H (2011) The research progress of sapovirus. Acta Veterinaria et Zootechnica Sinica, 42, 754-758. (in Chinese with English abstract) |
| [吴琼, 张艳, 郑海学, 殷宏 (2011) 札幌病毒研究进展. 畜牧兽医学报, 42, 754-758.] | |
| [29] |
Zhang FF, Luo SX, Gu J, Li ZQ, Li K, Yuan WF, Ye Y, Li H, Ding Z, Song DP, Tang YX (2019) Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in Southern China from 2012 to 2018. BMC Veterinary Research, 15, 470.
DOI PMID |
| [1] | Dandan Lang,Min Tang,Xin Zhou. Qualitative and quantitative molecular construction of plant-pollinator network: Application and prospective [J]. Biodiv Sci, 2018, 26(5): 445-456. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()