



Biodiv Sci ›› 2008, Vol. 16 ›› Issue (3): 263-270. DOI: 10.3724/SP.J.1003.2008.07362 cstr: 32101.14.SP.J.1003.2008.07362
• Original article • Previous Articles Next Articles
Juan Yan1,2, Haijia Chu1,2, Hengchang Wang1, Jianqiang Li1,*( )
)
Received:2007-12-03
Accepted:2008-02-15
Online:2008-05-20
Published:2008-05-20
Contact:
Jianqiang Li
About author:First author contact:**Contributed equally to this work
Juan Yan, Haijia Chu, Hengchang Wang, Jianqiang Li. Genetic structure and diversity of Medicago lupulina populations in northern and central China based on EST-SSRs markers[J]. Biodiv Sci, 2008, 16(3): 263-270.
| 居群 Population | 经纬度 Locality | 采样数目 Sample size |
|---|---|---|
| 新疆 Xinjiang | ||
| 1 喀什 Kashi | 39.44° N, 75.988° E | 23 |
| 2 伊犁 Yili | 43.88° N, 81.305° E | 20 |
| 3 那拉提 Nalati | 43.45° N, 83.283° E | 20 |
| 4 库尔勒 Korla | 41.75° N, 86.128° E | 17 |
| 5 奇台 Qitai | 43.70° N, 89.604° E | 23 |
| 6 巴里坤 Balikun | 43.58° N, 93.008° E | 20 |
| 内蒙古 Inner Mongolia | ||
| 7 海拉尔 Hailar | 48.90° N, 119.837° E | 18 |
| 8 白音敖包 Baiyinaobao | 43.53° N, 117.236° E | 21 |
| 9 兴和 Xinghe | 40.89° N, 113.885° E | 21 |
| 北京 Beijing | ||
| 10 东灵山 Dongling Mountain | 39.96° N, 115.439° E | 12 |
| 山西 Shanxi | ||
| 11 偏关 Pianguan | 39.44° N, 111.461° E | 22 |
| 陕西 Shaanxi | ||
| 12 鱼河 Yuhe | 37.98° N, 109.853° E | 23 |
| 13 南泥湾 Nanniwan | 36.32° N, 109.651° E | 20 |
| 14 西安 Xi’an | 34.28° N, 108.958° E | 27 |
| 甘肃 Gansu | ||
| 15 朱岔 Zhucha | 36.94° N, 102.589° E | 21 |
| 16 兴隆山 Xinglong Mountain 湖北 Hubei | 35.78° N, 104.048° E | 20 |
| 17 武汉 Wuhan | 30.55° N, 114.415° E | 26 |
| 总计 Total | 354 |
Table 1 Sampling sites and sample size of Medicago lupulinapopulations
| 居群 Population | 经纬度 Locality | 采样数目 Sample size |
|---|---|---|
| 新疆 Xinjiang | ||
| 1 喀什 Kashi | 39.44° N, 75.988° E | 23 |
| 2 伊犁 Yili | 43.88° N, 81.305° E | 20 |
| 3 那拉提 Nalati | 43.45° N, 83.283° E | 20 |
| 4 库尔勒 Korla | 41.75° N, 86.128° E | 17 |
| 5 奇台 Qitai | 43.70° N, 89.604° E | 23 |
| 6 巴里坤 Balikun | 43.58° N, 93.008° E | 20 |
| 内蒙古 Inner Mongolia | ||
| 7 海拉尔 Hailar | 48.90° N, 119.837° E | 18 |
| 8 白音敖包 Baiyinaobao | 43.53° N, 117.236° E | 21 |
| 9 兴和 Xinghe | 40.89° N, 113.885° E | 21 |
| 北京 Beijing | ||
| 10 东灵山 Dongling Mountain | 39.96° N, 115.439° E | 12 |
| 山西 Shanxi | ||
| 11 偏关 Pianguan | 39.44° N, 111.461° E | 22 |
| 陕西 Shaanxi | ||
| 12 鱼河 Yuhe | 37.98° N, 109.853° E | 23 |
| 13 南泥湾 Nanniwan | 36.32° N, 109.651° E | 20 |
| 14 西安 Xi’an | 34.28° N, 108.958° E | 27 |
| 甘肃 Gansu | ||
| 15 朱岔 Zhucha | 36.94° N, 102.589° E | 21 |
| 16 兴隆山 Xinglong Mountain 湖北 Hubei | 35.78° N, 104.048° E | 20 |
| 17 武汉 Wuhan | 30.55° N, 114.415° E | 26 |
| 总计 Total | 354 |
| 位点 Locus | 等位基因数 A | 预期杂合度 HE | 观察杂合度 HO | 遗传分化系数 FST | 基因流 Nm | 遗传分化系数RST |
|---|---|---|---|---|---|---|
| MTIC189 | 10 | 0.256 | 0.017 | 0.555 | 0.201 | 0.478 |
| MTIC432 | 5 | 0.177 | 0.034 | 0.643 | 0.139 | 0.58 |
| MTIC339 | 4 | 0.039 | 0.000 | 0.058 | 4.038 | -0.004 |
| MTIC345 | 8 | 0.320 | 0.085 | 0.463 | 0.290 | 0.447 |
| MTIC210 | 10 | 0.267 | 0.005 | 0.564 | 0.193 | 0.636 |
| MTIC14 | 6 | 0.180 | 0.000 | 0.604 | 0.164 | 0.703 |
| MTIC188 | 11 | 0.247 | 0.014 | 0.612 | 0.159 | 0.619 |
| MAA660870 | 5 | 0.157 | 0.003 | 0.605 | 0.163 | 0.508 |
| MtSSRNFAL05 | 7 | 0.266 | 0.017 | 0.649 | 0.135 | 0.528 |
| 平均 Mean | 7.333 | 0.212 | 0.020 | 0.528 | 0.224 | 0.499 |
Table 2 Genetic diversity in Medicago lupulina at nine EST-SSR loci
| 位点 Locus | 等位基因数 A | 预期杂合度 HE | 观察杂合度 HO | 遗传分化系数 FST | 基因流 Nm | 遗传分化系数RST |
|---|---|---|---|---|---|---|
| MTIC189 | 10 | 0.256 | 0.017 | 0.555 | 0.201 | 0.478 |
| MTIC432 | 5 | 0.177 | 0.034 | 0.643 | 0.139 | 0.58 |
| MTIC339 | 4 | 0.039 | 0.000 | 0.058 | 4.038 | -0.004 |
| MTIC345 | 8 | 0.320 | 0.085 | 0.463 | 0.290 | 0.447 |
| MTIC210 | 10 | 0.267 | 0.005 | 0.564 | 0.193 | 0.636 |
| MTIC14 | 6 | 0.180 | 0.000 | 0.604 | 0.164 | 0.703 |
| MTIC188 | 11 | 0.247 | 0.014 | 0.612 | 0.159 | 0.619 |
| MAA660870 | 5 | 0.157 | 0.003 | 0.605 | 0.163 | 0.508 |
| MtSSRNFAL05 | 7 | 0.266 | 0.017 | 0.649 | 0.135 | 0.528 |
| 平均 Mean | 7.333 | 0.212 | 0.020 | 0.528 | 0.224 | 0.499 |
| 居群 Population | 平均等位基因数na | 有效等位基因数ne | 观察杂合度 Ho | 预期杂合度 HE | 近交系数 F | 自交率 s | 多态率 PPL (%) | 私有基因数 Private alleles |
|---|---|---|---|---|---|---|---|---|
| 1 喀什 Kashi | 2.444 | 1.746 | 0.019 | 0.346 | 0.947 | 0.973 | 66.7 | 0 |
| 2 伊犁 Yili | 3.556 | 1.717 | 0.056 | 0.342 | 0.845 | 0.916 | 100 | 5 |
| 3 那拉提 Nalati | 2.667 | 1.868 | 0.028 | 0.388 | 0.932 | 0.965 | 88.9 | 1 |
| 4 库尔勒 Korla | 2.222 | 1.779 | 0.007 | 0.348 | 0.982 | 0.991 | 77.8 | 1 |
| 5 奇台 Qitai | 2.111 | 1.380 | 0.000 | 0.214 | 1.000 | 1.000 | 77.8 | 0 |
| 6 巴里坤 Balikun | 1.667 | 1.196 | 0.034 | 0.118 | 0.725 | 0.841 | 33.3 | 0 |
| 7 海拉尔 Hailar | 2.111 | 1.137 | 0.025 | 0.107 | 0.781 | 0.877 | 66.7 | 1 |
| 8 白音敖包 Baiyinaobao | 1.778 | 1.196 | 0.011 | 0.145 | 0.93 | 0.964 | 66.7 | 0 |
| 9 兴和 Xinghe | 1.889 | 1.223 | 0.011 | 0.153 | 0.931 | 0.964 | 77.8 | 0 |
| 10 东灵山 Dongling Mountain | 2.111 | 1.467 | 0.009 | 0.270 | 0.968 | 0.984 | 77.8 | 0 |
| 11 偏关 Pianguan | 2.000 | 1.183 | 0.035 | 0.137 | 0.752 | 0.858 | 88.9 | 0 |
| 12 鱼河 Yuhe | 2.333 | 1.294 | 0.034 | 0.188 | 0.824 | 0.904 | 66.7 | 1 |
| 13 南泥湾 Nanniwan | 1.667 | 1.246 | 0.000 | 0.152 | 1.000 | 1.000 | 55.6 | 1 |
| 14 西安 Xi’an | 1.444 | 1.049 | 0.012 | 0.042 | 0.716 | 0.835 | 33.3 | 0 |
| 15 朱岔 Zhucha | 3.556 | 1.448 | 0.042 | 0.252 | 0.840 | 0.913 | 77.8 | 8 |
| 16 兴隆山 Xinglongshan | 2.333 | 1.576 | 0.000 | 0.302 | 1.000 | 1.000 | 66.7 | 0 |
| 17 武汉 Wuhan | 2.333 | 1.122 | 0.009 | 0.104 | 0.921 | 0.959 | 100 | 3 |
| 平均 Mean | 2.248 | 1.390 | 0.020 | 0.212 | 0.888 | 0.938 | 71.9 | 1 |
Table 3 Genetic variability estimates for 17 Medicago lupulinapopulations
| 居群 Population | 平均等位基因数na | 有效等位基因数ne | 观察杂合度 Ho | 预期杂合度 HE | 近交系数 F | 自交率 s | 多态率 PPL (%) | 私有基因数 Private alleles |
|---|---|---|---|---|---|---|---|---|
| 1 喀什 Kashi | 2.444 | 1.746 | 0.019 | 0.346 | 0.947 | 0.973 | 66.7 | 0 |
| 2 伊犁 Yili | 3.556 | 1.717 | 0.056 | 0.342 | 0.845 | 0.916 | 100 | 5 |
| 3 那拉提 Nalati | 2.667 | 1.868 | 0.028 | 0.388 | 0.932 | 0.965 | 88.9 | 1 |
| 4 库尔勒 Korla | 2.222 | 1.779 | 0.007 | 0.348 | 0.982 | 0.991 | 77.8 | 1 |
| 5 奇台 Qitai | 2.111 | 1.380 | 0.000 | 0.214 | 1.000 | 1.000 | 77.8 | 0 |
| 6 巴里坤 Balikun | 1.667 | 1.196 | 0.034 | 0.118 | 0.725 | 0.841 | 33.3 | 0 |
| 7 海拉尔 Hailar | 2.111 | 1.137 | 0.025 | 0.107 | 0.781 | 0.877 | 66.7 | 1 |
| 8 白音敖包 Baiyinaobao | 1.778 | 1.196 | 0.011 | 0.145 | 0.93 | 0.964 | 66.7 | 0 |
| 9 兴和 Xinghe | 1.889 | 1.223 | 0.011 | 0.153 | 0.931 | 0.964 | 77.8 | 0 |
| 10 东灵山 Dongling Mountain | 2.111 | 1.467 | 0.009 | 0.270 | 0.968 | 0.984 | 77.8 | 0 |
| 11 偏关 Pianguan | 2.000 | 1.183 | 0.035 | 0.137 | 0.752 | 0.858 | 88.9 | 0 |
| 12 鱼河 Yuhe | 2.333 | 1.294 | 0.034 | 0.188 | 0.824 | 0.904 | 66.7 | 1 |
| 13 南泥湾 Nanniwan | 1.667 | 1.246 | 0.000 | 0.152 | 1.000 | 1.000 | 55.6 | 1 |
| 14 西安 Xi’an | 1.444 | 1.049 | 0.012 | 0.042 | 0.716 | 0.835 | 33.3 | 0 |
| 15 朱岔 Zhucha | 3.556 | 1.448 | 0.042 | 0.252 | 0.840 | 0.913 | 77.8 | 8 |
| 16 兴隆山 Xinglongshan | 2.333 | 1.576 | 0.000 | 0.302 | 1.000 | 1.000 | 66.7 | 0 |
| 17 武汉 Wuhan | 2.333 | 1.122 | 0.009 | 0.104 | 0.921 | 0.959 | 100 | 3 |
| 平均 Mean | 2.248 | 1.390 | 0.020 | 0.212 | 0.888 | 0.938 | 71.9 | 1 |
| 变异来源 Source of variation | 自由度 d. f. | 离差平方和(SSD) Sum of squares | 方差分量 Variance components | 方差分量比例(%) Percentage of variation | P |
|---|---|---|---|---|---|
| 居群间 Among populations | 16 | 951.886 | Va = 1.38653 | 59.02 | P<0.001 |
| 居群内个体间 Within populations | 337 | 619.494 | Vb = 0.87535 | 37.26 | P<0.001 |
| 个体内 Within individuals | 354 | 31.000 | Vc = 0.08757 | 3.73 | P<0.001 |
| 总计 Total | 707 | 1602.380 | 2.34945 |
Table 4 Molecular variance (AMOVA) analysis based on EST-SSR markers for Medicago lupulina
| 变异来源 Source of variation | 自由度 d. f. | 离差平方和(SSD) Sum of squares | 方差分量 Variance components | 方差分量比例(%) Percentage of variation | P |
|---|---|---|---|---|---|
| 居群间 Among populations | 16 | 951.886 | Va = 1.38653 | 59.02 | P<0.001 |
| 居群内个体间 Within populations | 337 | 619.494 | Vb = 0.87535 | 37.26 | P<0.001 |
| 个体内 Within individuals | 354 | 31.000 | Vc = 0.08757 | 3.73 | P<0.001 |
| 总计 Total | 707 | 1602.380 | 2.34945 |
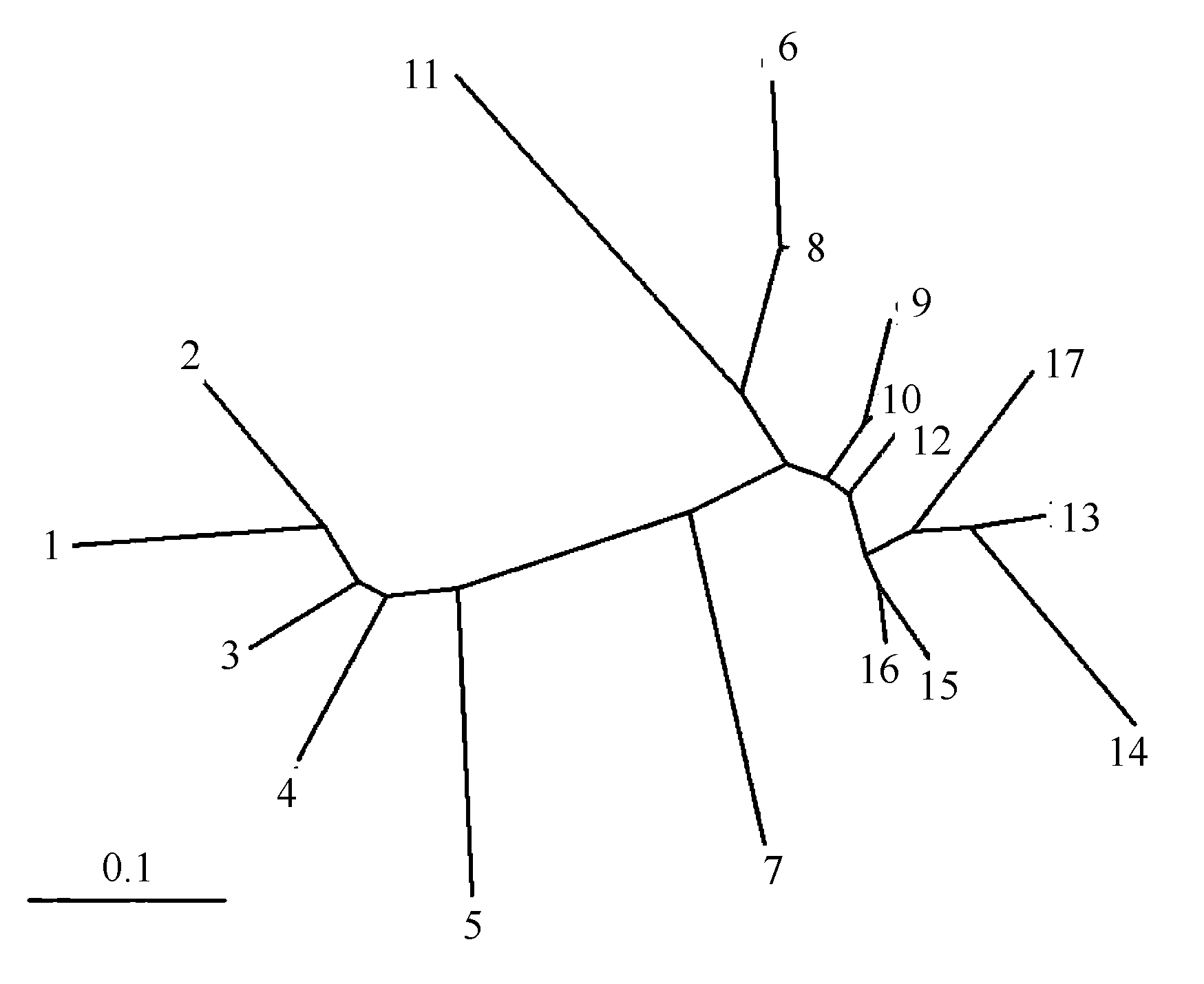
Fig. 4 Unrooted neighbour-joining tree showing relationships between the 17 Medicago lupulina populations. Population codes are the same as in Table 1.
| [1] | Bohonak AJ (2002) IBD (Isolation by Distance): a program for analyses of isolation by distance. Journal of Heredity, 93,153-154. |
| [2] | Bonnin I, Ronfort J, Wozniak F, Olivier I (2001) Spatial effects and rare outcrossing events in Medicago truncatula (Fabaceae) . Molecular Ecology, 10,1371-1383. |
| [3] | Bouck A, Vision T (2007) The molecular ecologist’s guide to expressed sequence tags. Molecular Ecology, 16,907-924. |
| [4] | Cao ZZ (曹致中), Feng YQ (冯毓琴), Ma HL (马晖玲), Liu XN (柳小妮), Zhou YL (周玉雷), Xu ZM (徐智明) (2003) Medicago lupulina—beautiful water-saving and easily-maintained turf legume . Pratacultural Science (草业科学), 20(4),58-60. (in Chinese with English abstract) |
| [5] | Chabane K, Ablett GA, Cordeiro GM, Valkoun J, Henry RJ (2005) EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genetic Resources and Crop Evolution, 52,903-909. |
| [6] | Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice ( Oryza sativa L.) . Theoretical and Applied Genetics, 100,713-722. |
| [7] | Clapham AR, Tutin TG, Warburg EF (1962) Flora of the British Isles. Cambridge University Press,Cambridge, UK. |
| [8] | Clauss MJ, Mitchell-Olds T (2006) Population genetic structure of Arabidopsis lyrata in Europe . Molecular Ecology, 15,2753-2766. |
| [9] | Crow JK, Kimura M (1970) An Introduction to Population Genetic Theory. Harper and Row, New York. |
| [10] | Diwan N, Bouton JH, Kochert G, Cregan PB (2000) Mapping of simple sequence repeat (SSR) DNA markers in diploid and tetraploid alfalfa. Theoretical and Applied Genetics, 101,165-172. |
| [11] | Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19,11-15. |
| [12] | Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity, 99,125-132. |
| [13] | Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity, 72,250-259. |
| [14] | Eujayl I, Sledge MK, Wang L, May GD, Chekhovskiy K, Zwonitzer J, Mian MAR (2004) Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. Theoretical and Applied Genetics, 108,414-422. |
| [15] | Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14,2611-2620. |
| [16] | Flajoulot S, Ronfort J, Baudouin P, Barre P, Huguet T, Huyghe C, Julier B (2005) Genetic diversity among alfalfa ( Medicago sativa) cultivars coming from a breeding program, using SSR markers . Theoretical and Applied Genetics, 111,1420-1429. |
| [17] | Fowler DP (1965) Effects of inbreeding in red pine, Pinus resinosa Ait. Ⅱ. Pollination studies . Silvae Genetica, 14,12-23. |
| [18] | Goudet J (2001) FSTAT, A Program to Estimate and Test Gene Diversities and Fixation Indices, version 2. 9.3. http://www.unil.ch/izea/softwares/fstat.html. |
| [19] | Govindaraju DR (1988) Relationship between dispersal ability and levels of gene flow in plants. Oikos, 52,31-35. |
| [20] |
Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Molecular Genetics and Genomics, 270,315-323.
URL PMID |
| [21] | Hamrick JL, Godt MJW (1996) Effects of life-history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences, 351,1291-1298. |
| [22] | Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C (2003) Construction of two genetic linkage maps in cultivated tetraploid alfalfa ( Medicago sativa) using microsatellite and AFLP markers . BMC Plant Biology, 3,9. |
| [23] |
Kuroda Y, Kaga A, Tomooka N, Vaughan A (2006) Population genetic structure of Japanese wild soybean ( Glycine soja) based on microsatellite variation . Molecular Ecology, 15,959-974.
URL PMID |
| [24] | Lammerink J (1968) Genetic variability in commencement of flowering in Medicago lupulina L. in the south island of New Zealand . New Zealand Journal of Botany, 6,33-42. |
| [25] | Langella O (2000) Populations 1.2: Population Genetic Software (Individuals or Population Distance, Phylogenetic Trees). http://bioinformatics.org/~tryphon/popula- tions/. |
| [26] | Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics, 15,65-95. |
| [27] |
Mable BK, Adam A (2007) Patterns of genetic diversity in outcrossing and selfing populations of Arabidopsis lyrata. Molecular Ecology, 16,3565-3580.
URL PMID |
| [28] | Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Research, 27,209-220. |
| [29] |
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution, 19,153-170.
DOI URL PMID |
| [30] | Moyle LC (2006) Correlates of genetic differentiation and isolation by distance in 17 congeneric Silene species . Molecular Ecology, 15,1067-1081. |
| [31] |
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology, 13,1143-1155.
DOI URL PMID |
| [32] |
Pashley CH, Ellis JR, McCauley DE, Burke JM (2006) EST databases as a source for molecular markers: lessons from Helianthus. Journal of Heredity, 97,381-388.
URL PMID |
| [33] | Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences, 12,357-358. |
| [34] | Pavone LV, Reader RJ (1985) Effect of microtopography on the survival and reproduction of Medicago lupulina . The Journal of Ecology, 73,685-694. |
| [35] | Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A (1998) Cross species amplification of soybean ( Glycine max) simple sequence repeat (SSRs) within the genus and other legume genera: implication for transferability of SSRs in plants . Molecular Biology and Evolution, 15,1275-1287. |
| [36] | Peakall R, Smouse PE (2006) GenALEx6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6,288-295. |
| [37] | Pritchard JK, Stephans M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155,945-959. |
| [38] | Pritchard JK, Wen XQ, Fslush D (2007) STRUCTURE: Documentation for Structure Software: version 2.2. http://pritch.bsd.uchicago.edu/software. |
| [39] | Rice WR (1989) Analyzing tables of statistical tests. Evolution, 43,223-225. |
| [40] | Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance . Genetics, 145,1219-1228. |
| [41] | Sanguinetti CJ, Dias NE, Simpson G (1994) Rapid silver staining and recovery of PCR products separated on polyaerylamide gels. Biotechniques, 17,915-919. |
| [42] | Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN ver. 2.000: A Software for Population Genetics Data Analysis. Genetics and Biochemistry Laboratory, University of Geneva, Switzerland. |
| [43] | Sidhu SS (1971) Some Aspects of the Ecology of Black Medick (Medicago lupulina L.). PhD dissertation, University of Western Ontario., . |
| [44] | Slatkin M, Barton NH (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution, 43,1349-1368. |
| [45] | StenØien HK, Fenster CB, Tonteri A, Savolainen O (2005) Genetic variability in natural populations of Arabidopsis thaliana in northern Europe . Molecular Ecology, 14,137-148. |
| [46] | Turkington RA, Cavers PB (1979) The biology of Canadian weeds. 33. Medicago lupulina L. Canadian Journal of Plant Science, 59,99-110. |
| [47] | Wei Z (韦直), Huang YZ (黄以之) (1998) Leguminosae. In:Flora Reipublicae Popularis Sinicae (中国植物志), Tomus 42, pp.314-316. Science Press, Beijing.. (in Chinese) |
| [48] | Wright S (1965) The interpretations of population structure by F-statistics with special regards to systems of mating . Evolution, 19,395-420. |
| [49] | Zhu BC (朱邦长), He SJ (何胜江), Zhang CQ (张川黔), Song GX (宋高翔) (1996) Guizhou natural legume herbage—introduction and domestication of black medick (Medicago lupulina L.). Guizhou Agricultural Sciences (贵州农业科学), 24(4),36-40. (in Chinese with English abstract) |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [3] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [4] | Xianglin Yang, Caiyun Zhao, Junsheng Li, Fangfang Chong, Wenjin Li. Invasive plant species lead to a more clustered community phylogenetic structure: An analysis of herbaceous plants in Guangxi’s national nature reserves [J]. Biodiv Sci, 2024, 32(11): 24175-. |
| [5] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [6] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [7] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [8] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [9] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [10] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [11] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [12] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [13] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [14] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [15] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2026 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn