



Biodiv Sci ›› 2025, Vol. 33 ›› Issue (7): 25119. DOI: 10.17520/biods.2025119 cstr: 32101.14.biods.2025119
• Original Papers: Microbial Diversity • Previous Articles Next Articles
Xinbo Hou1( ), Xiuhai Zhao1, Huaijiang He2, Chunyu Zhang1, Juan Wang3, Xueying Ren1, Xinna Zhang1,*(
), Xiuhai Zhao1, Huaijiang He2, Chunyu Zhang1, Juan Wang3, Xueying Ren1, Xinna Zhang1,*( )(
)( )
)
Received:2025-04-01
Accepted:2025-06-18
Online:2025-07-20
Published:2025-08-27
Contact:
*E-mail: zhangxinna@bjfu.edu.cn
Supported by:Xinbo Hou, Xiuhai Zhao, Huaijiang He, Chunyu Zhang, Juan Wang, Xueying Ren, Xinna Zhang. Responses of rhizosphere and non-rhizosphere microbial communities to the soil carbon and nitrogen in Quercus mongolica pure forest[J]. Biodiv Sci, 2025, 33(7): 25119.
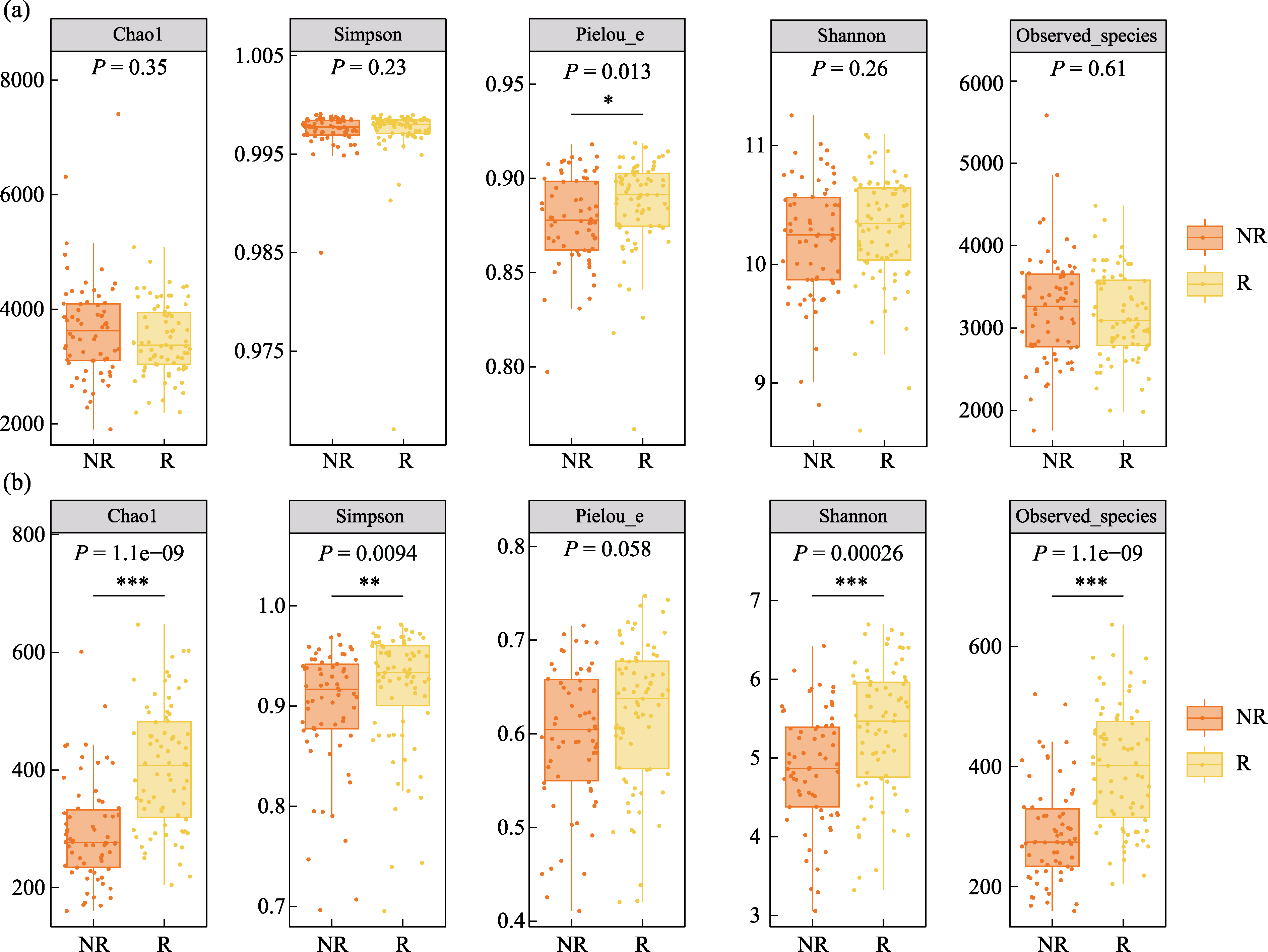
Fig. 2 α diversity indices of bacterial (a) and fungal (b) communities in rhizosphere (R) and non-rhizosphere (NR) soils. Chao1, Chao1 richness index; Simpson, Simpson diversity index; Pielou_e, Pielou evenness index; Shannon, Shannon diversity index; Observed_species, Observed species richness index. * P < 0.05; ** P < 0.01; *** P < 0.001.
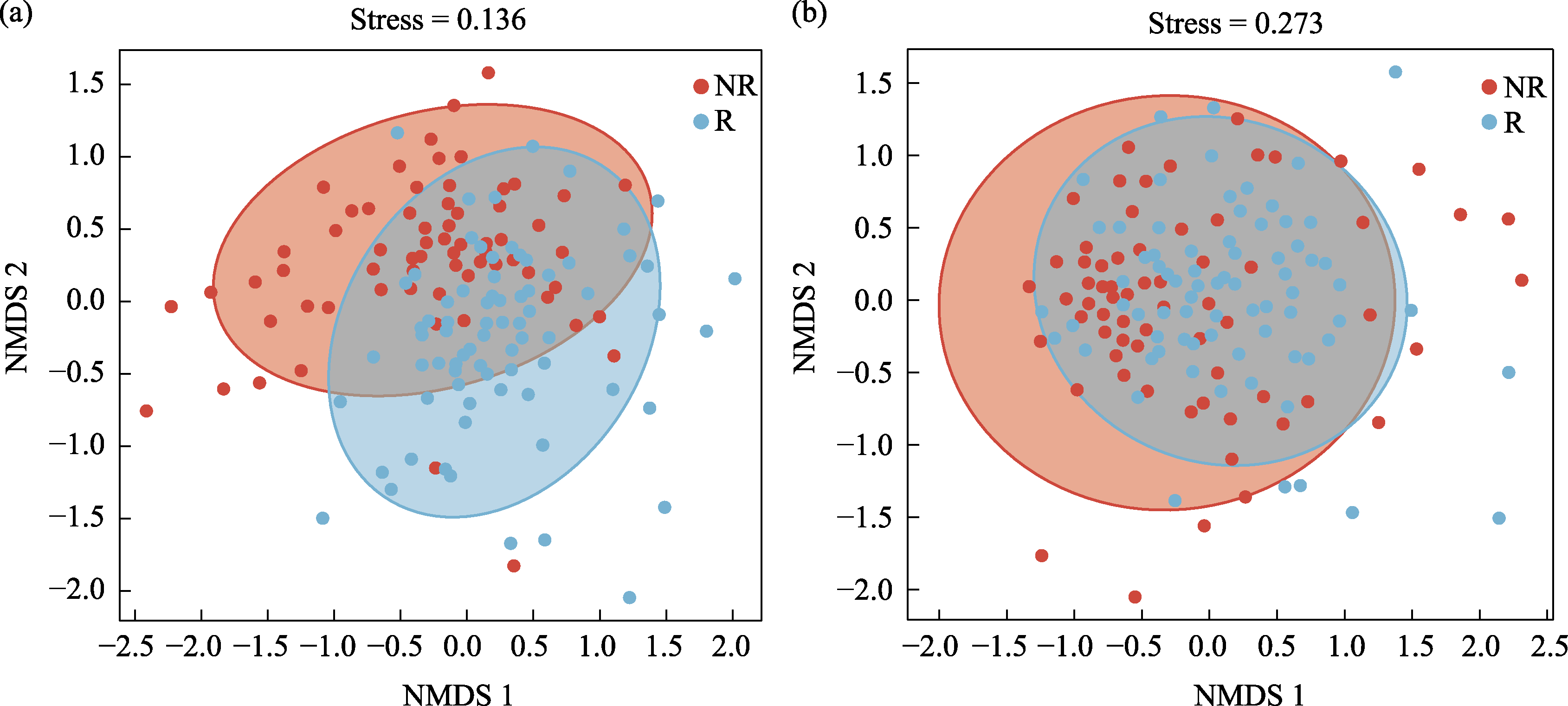
Fig. 3 Non-metric multidimensional scaling (NMDS) analysis of bacterial (a) and fungal (b) communities in rhizosphere (R) and non-rhizosphere (NR) soils based on Bray-Curtis distance
| 样本 Sample | 基于Bray-Curtis距离的F统计值 F-statistic value based on Bray-Curtis distance (P) | |||
|---|---|---|---|---|
| R1 | R2 | R3 | ||
| 细菌 Bacteria | NR1 | 4.78 (0.001) | 7.23 (0.001) | 7.19 (0.001) |
| NR2 | 2.62 (0.001) | 3.63 (0.001) | 4.05 (0.001) | |
| NR3 | 3.56 (0.001) | 3.58 (0.001) | 1.80 (0.002) | |
| 真菌 Fungi | NR1 | 2.47 (0.001) | 4.95 (0.001) | 5.29 (0.001) |
| NR2 | 2.93 (0.001) | 2.28 (0.001) | 2.43 (0.001) | |
| NR3 | 2.43 (0.001) | 2.58 (0.001) | 1.23 (0.093) | |
Table 1 Analysis of intergroup differences in soil microbial communities between rhizosphere (R) and non-rhizosphere (NR) soils (PERMANOVA test based on Bray-Curtis distance). R1-3 and NR1-3 represent rhizosphere and non-rhizosphere soils from sampling sites 1-3, respectively.
| 样本 Sample | 基于Bray-Curtis距离的F统计值 F-statistic value based on Bray-Curtis distance (P) | |||
|---|---|---|---|---|
| R1 | R2 | R3 | ||
| 细菌 Bacteria | NR1 | 4.78 (0.001) | 7.23 (0.001) | 7.19 (0.001) |
| NR2 | 2.62 (0.001) | 3.63 (0.001) | 4.05 (0.001) | |
| NR3 | 3.56 (0.001) | 3.58 (0.001) | 1.80 (0.002) | |
| 真菌 Fungi | NR1 | 2.47 (0.001) | 4.95 (0.001) | 5.29 (0.001) |
| NR2 | 2.93 (0.001) | 2.28 (0.001) | 2.43 (0.001) | |
| NR3 | 2.43 (0.001) | 2.58 (0.001) | 1.23 (0.093) | |
| 属 Genus | 功能描述 Functional description | 相对丰度 Relative abundance | 参考文献 References | ||
|---|---|---|---|---|---|
| NR | R | ||||
| 细菌 Bacteria | Candidatus Udaeobacter | 有机污染物降解与碳代谢 Degradation of organic pollutants and carbon metabolism | 0.043 ± 0.022A | 0.029 ± 0.019B | 童彤等, |
| Burkholderia-Caballeronia- Paraburkholderia | 有机污染物降解 Degradation of organic pollutants | 0.016 ± 0.014A | 0.036 ± 0.033B | 李霏等, | |
| Candidatus Solibacter | 抗重金属 Resistant to heavy metals | 0.020 ± 0.005a | 0.018 ± 0.005b | Puthusseri et al, | |
| Rokubacteriales | 氮呼吸 Nitrogen respiration | 0.026 ± 0.026A | 0.008 ± 0.010B | Reis et al, | |
| Bradyrhizobium | 共生和固氮 Symbiosis and nitrogen fixation | 0.014 ± 0.005a | 0.016 ± 0.005b | 汪其同等, | |
| Acidothermus | 纤维素分解 Degrade cellulose | 0.010 ± 0.006A | 0.018 ± 0.009B | Rezaei et al, | |
| Acidibacter | 蛋白质分解 Degrade proteins | 0.011 ± 0.004A | 0.015 ± 0.005B | 何庆海等, | |
| Mycobacterium | 致病菌 Pathogenic bacteria | 0.010 ± 0.005A | 0.014 ± 0.005B | Brown-Elliott & Wallace, | |
| Granulicella | 聚合物降解 Polymer degradation | 0.006 ± 0.006A | 0.011 ± 0.008B | Oshkin et al, | |
| Haliangium | 抗植物病原真菌 Resistant to plant pathogenic fungi | 0.006 ± 0.003A | 0.009 ± 0.006B | Kundim et al, | |
| 真菌 Fungi | Trichoderma | 菌寄生 Mycoparasitism | 0.029 ± 0.038 | 0.069 ± 0.069 | Khan et al, |
| Geminibasidium | 共生 Symbiosis | 0.053 ± 0.072a | 0.028 ± 0.044b | Ren et al, | |
| Lactarius | 抗肿瘤 Anti-tumor | 0.010 ± 0.027a | 0.026 ± 0.052b | 王和等, | |
| Saitozyma | 纤维素分解 Degrade cellulose | 0.010 ± 0.027a | 0.026 ± 0.052b | Aliyu et al, | |
| Cortinarius | 共生 Symbiosis | 0.001 ± 0.007a | 0.016 ± 0.056b | Soop et al, | |
Table 2 Functional description of some genera with significant changes in the relative abundance of rhizosphere (R) and non-rhizosphere (NR) soil bacteria and fungi
| 属 Genus | 功能描述 Functional description | 相对丰度 Relative abundance | 参考文献 References | ||
|---|---|---|---|---|---|
| NR | R | ||||
| 细菌 Bacteria | Candidatus Udaeobacter | 有机污染物降解与碳代谢 Degradation of organic pollutants and carbon metabolism | 0.043 ± 0.022A | 0.029 ± 0.019B | 童彤等, |
| Burkholderia-Caballeronia- Paraburkholderia | 有机污染物降解 Degradation of organic pollutants | 0.016 ± 0.014A | 0.036 ± 0.033B | 李霏等, | |
| Candidatus Solibacter | 抗重金属 Resistant to heavy metals | 0.020 ± 0.005a | 0.018 ± 0.005b | Puthusseri et al, | |
| Rokubacteriales | 氮呼吸 Nitrogen respiration | 0.026 ± 0.026A | 0.008 ± 0.010B | Reis et al, | |
| Bradyrhizobium | 共生和固氮 Symbiosis and nitrogen fixation | 0.014 ± 0.005a | 0.016 ± 0.005b | 汪其同等, | |
| Acidothermus | 纤维素分解 Degrade cellulose | 0.010 ± 0.006A | 0.018 ± 0.009B | Rezaei et al, | |
| Acidibacter | 蛋白质分解 Degrade proteins | 0.011 ± 0.004A | 0.015 ± 0.005B | 何庆海等, | |
| Mycobacterium | 致病菌 Pathogenic bacteria | 0.010 ± 0.005A | 0.014 ± 0.005B | Brown-Elliott & Wallace, | |
| Granulicella | 聚合物降解 Polymer degradation | 0.006 ± 0.006A | 0.011 ± 0.008B | Oshkin et al, | |
| Haliangium | 抗植物病原真菌 Resistant to plant pathogenic fungi | 0.006 ± 0.003A | 0.009 ± 0.006B | Kundim et al, | |
| 真菌 Fungi | Trichoderma | 菌寄生 Mycoparasitism | 0.029 ± 0.038 | 0.069 ± 0.069 | Khan et al, |
| Geminibasidium | 共生 Symbiosis | 0.053 ± 0.072a | 0.028 ± 0.044b | Ren et al, | |
| Lactarius | 抗肿瘤 Anti-tumor | 0.010 ± 0.027a | 0.026 ± 0.052b | 王和等, | |
| Saitozyma | 纤维素分解 Degrade cellulose | 0.010 ± 0.027a | 0.026 ± 0.052b | Aliyu et al, | |
| Cortinarius | 共生 Symbiosis | 0.001 ± 0.007a | 0.016 ± 0.056b | Soop et al, | |
| 土壤类型 Soil types | 全碳 Total carbon (TC, g/kg) | 全氮 Total nitrogen (TN, g/kg) | 土壤有机碳 Soil organic carbon (SOC, g/kg) | C : N |
|---|---|---|---|---|
| 非根际土壤 Non-rhizosphere soil (NR) | 4.40 ± 1.49A | 0.30 ± 0.09A | 1.43 ± 3.49A | 0.28 ± 0.36a |
| 根际土壤 Rhizosphere soil (R) | 7.55 ± 3.22B | 0.50 ± 0.22B | 3.79 ± 2.01B | 0.56 ± 0.63b |
Table 3 Stoichiometric characteristics of soil carbon and nitrogen
| 土壤类型 Soil types | 全碳 Total carbon (TC, g/kg) | 全氮 Total nitrogen (TN, g/kg) | 土壤有机碳 Soil organic carbon (SOC, g/kg) | C : N |
|---|---|---|---|---|
| 非根际土壤 Non-rhizosphere soil (NR) | 4.40 ± 1.49A | 0.30 ± 0.09A | 1.43 ± 3.49A | 0.28 ± 0.36a |
| 根际土壤 Rhizosphere soil (R) | 7.55 ± 3.22B | 0.50 ± 0.22B | 3.79 ± 2.01B | 0.56 ± 0.63b |
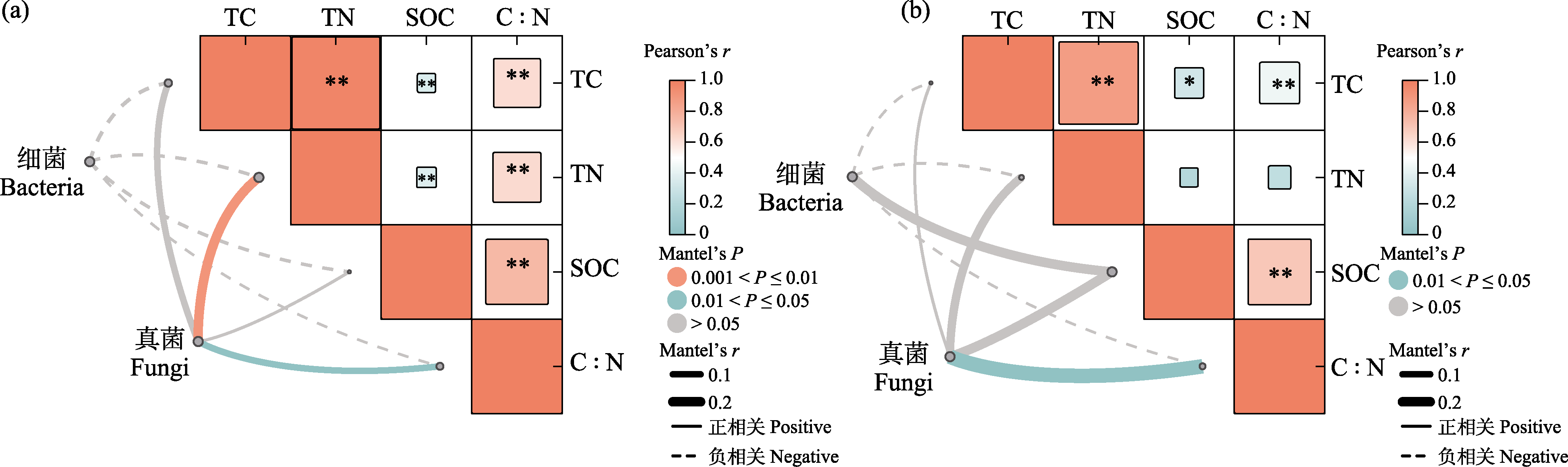
Fig. 4 Mantel test correlation analysis between microbial communities and soil carbon-nitrogen factors in rhizosphere (a) and non-rhizosphere (b) soils. Color gradient within grids represents Pearson’s correlation coefficient, * P < 0.05, ** P < 0.01; the color of connecting lines indicates statistical significance, the width represents the Mantel’s r statistic, solid lines indicate positive correlations, dashed lines indicate negative correlations, orange indicates highly significant correlations (P < 0.01), and green indicates significant correlations (P < 0.05). TC, Total carbon; TN, Total nitrogen; SOC, Soil organic carbon.
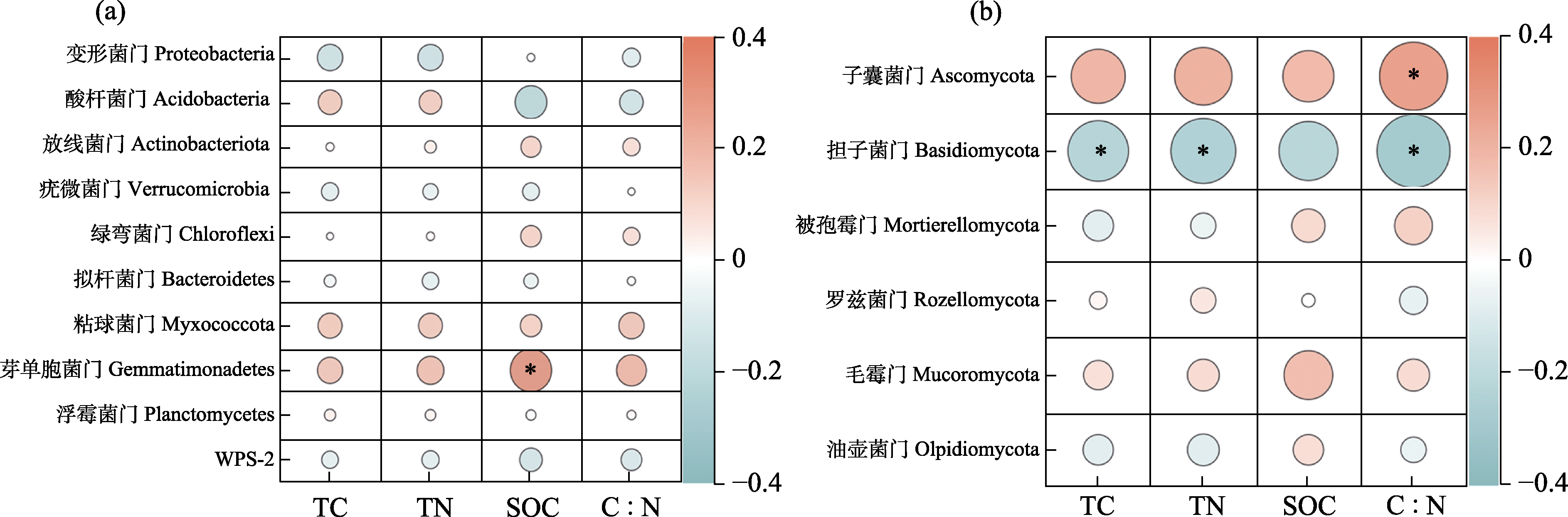
Fig. 5 Pearson correlation analysis of dominant bacterial (a) and fungal (b) phyla in rhizosphere soil as related to soil carbon and nitrogen factors. The color bar scale represents the Pearson correlation coefficient (r) value. Orange represents a positive correlation, and green represents a negative correlation; The shade of color indicates the strength of the correlation; * P < 0.05. TC, Total carbon; TN, Total nitrogen; SOC, Soil organic carbon.
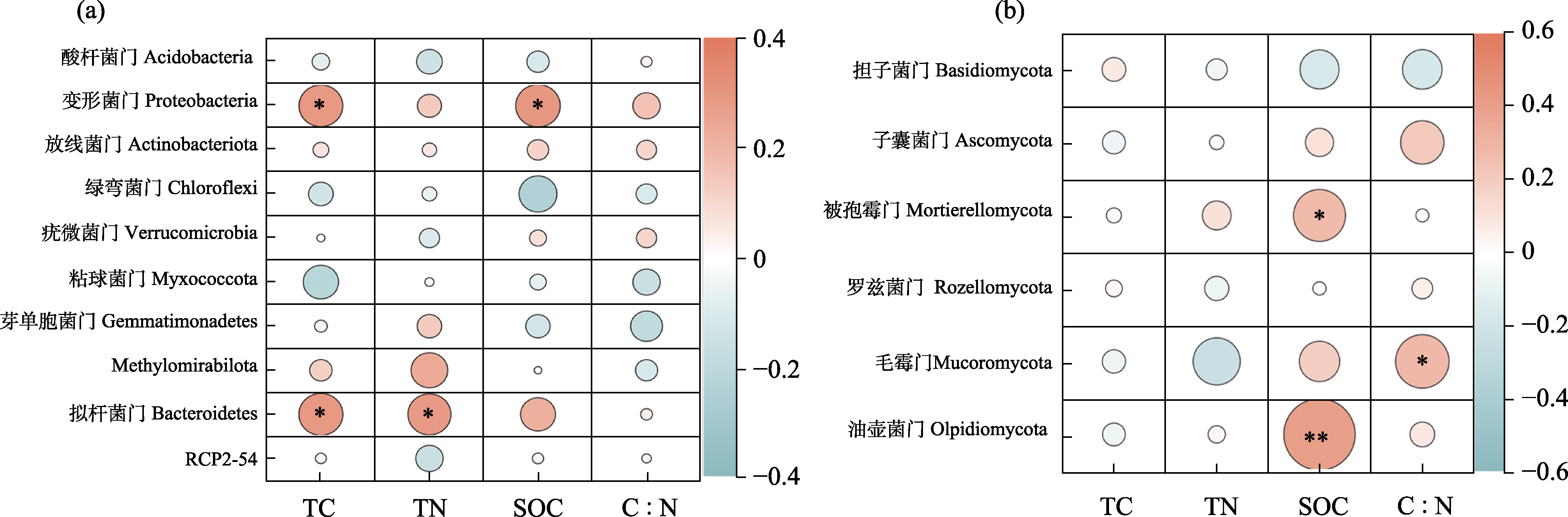
Fig. 6 Pearson correlation analysis of dominant bacterial (a) and fungal (b) phyla in non-rhizosphere soil as related to soil carbon and nitrogen factors. The color bar scale represents the Pearson correlation coefficient (r) value. Orange represents a positive correlation, and green represents a negative correlation; The shade of color indicates the strength of the correlation; * P < 0.05, ** P < 0.01. TC, Total carbon; TN, Total nitrogen; SOC, Soil organic carbon.
| 土壤类型 Soil types | 碳氮指标 Carbon and nitrogen indicators | Shannon多样性 指数 Shannon diversity index | Simpson多样性 指数 Simpson diversity index | Pielou均匀度 指数 Pielou evenness index | Chao1丰富度 指数 Chao1 richness index | Observed species丰富度指数 Observed species richness index | |
|---|---|---|---|---|---|---|---|
| 细菌 Bacteria | NR | TC | 0.098 | 0.070 | 0.110 | -0.039 | 0.037 |
| SOC | 0.084 | 0.042 | 0.105 | -0.033 | 0.024 | ||
| TN | 0.238 | 0.144 | 0.259* | 0.051 | 0.145 | ||
| C : N | -0.090 | -0.099 | -0.071 | -0.133 | -0.112 | ||
| R | TC | 0.142 | 0.121 | 0.160 | 0.068 | 0.096 | |
| SOC | 0.303** | 0.125 | 0.272* | 0.252* | 0.302** | ||
| TN | 0.149 | 0.129 | 0.163 | 0.084 | 0.108 | ||
| C : N | 0.196 | 0.110 | 0.222 | 0.065 | 0.127 | ||
| 真菌 Fungi | NR | TC | 0.115 | 0.047 | -0.005 | 0.317* | 0.329* |
| SOC | 0.169 | 0.149 | 0.116 | 0.199 | 0.207 | ||
| TN | 0.165 | 0.073 | 0.039 | 0.360** | 0.378** | ||
| C : N | 0.115 | 0.095 | 0.103 | 0.065 | 0.068 | ||
| R | TC | 0.092 | 0.122 | 0.105 | 0.034 | 0.031 | |
| SOC | 0.102 | 0.181 | 0.168 | -0.134 | -0.125 | ||
| TN | 0.102 | 0.109 | 0.110 | 0.050 | 0.047 | ||
| C : N | 0.021 | 0.121 | 0.073 | -0.137 | -0.134 |
Table 4 Correlation between rhizosphere (R) and non-rhizosphere (NR) soil microbial α diversity index and soil C and N indices
| 土壤类型 Soil types | 碳氮指标 Carbon and nitrogen indicators | Shannon多样性 指数 Shannon diversity index | Simpson多样性 指数 Simpson diversity index | Pielou均匀度 指数 Pielou evenness index | Chao1丰富度 指数 Chao1 richness index | Observed species丰富度指数 Observed species richness index | |
|---|---|---|---|---|---|---|---|
| 细菌 Bacteria | NR | TC | 0.098 | 0.070 | 0.110 | -0.039 | 0.037 |
| SOC | 0.084 | 0.042 | 0.105 | -0.033 | 0.024 | ||
| TN | 0.238 | 0.144 | 0.259* | 0.051 | 0.145 | ||
| C : N | -0.090 | -0.099 | -0.071 | -0.133 | -0.112 | ||
| R | TC | 0.142 | 0.121 | 0.160 | 0.068 | 0.096 | |
| SOC | 0.303** | 0.125 | 0.272* | 0.252* | 0.302** | ||
| TN | 0.149 | 0.129 | 0.163 | 0.084 | 0.108 | ||
| C : N | 0.196 | 0.110 | 0.222 | 0.065 | 0.127 | ||
| 真菌 Fungi | NR | TC | 0.115 | 0.047 | -0.005 | 0.317* | 0.329* |
| SOC | 0.169 | 0.149 | 0.116 | 0.199 | 0.207 | ||
| TN | 0.165 | 0.073 | 0.039 | 0.360** | 0.378** | ||
| C : N | 0.115 | 0.095 | 0.103 | 0.065 | 0.068 | ||
| R | TC | 0.092 | 0.122 | 0.105 | 0.034 | 0.031 | |
| SOC | 0.102 | 0.181 | 0.168 | -0.134 | -0.125 | ||
| TN | 0.102 | 0.109 | 0.110 | 0.050 | 0.047 | ||
| C : N | 0.021 | 0.121 | 0.073 | -0.137 | -0.134 |
| [1] | Ai C, Liang GQ, Sun JW, Wang XB, He P, Zhou W, He XH (2015) Reduced dependence of rhizosphere microbiome on plant-derived carbon in 32-year long-term inorganic and organic fertilized soils. Soil Biology and Biochemistry, 80, 70-78. |
| [2] |
Aleklett K, Ohlsson P, Bengtsson M, Hammer EC (2021) Fungal foraging behaviour and hyphal space exploration in micro-structured Soil Chips. ISME Journal, 15, 1782-1793.
DOI PMID |
| [3] | Aliyu H, Gorte O, Neumann A, Ochsenreither K (2021) Global transcriptome profile of the oleaginous yeast Saitozyma podzolica DSM 27192 cultivated in glucose and xylose. Journal of Fungi, 7, 758. |
| [4] | Ban MJ, Zhao N, Peng SL, Ge ZW, Xing W, Li NN, Mao LF (2024) Meta-analysis of the contributions of microbial necromass carbon on soil organic carbon in arbuscular mycorrhizae-dominated and ectomycorrhizae-dominated forests. Acta Ecologica Sinica, 44, 11254-11264. (in Chinese with English abstract) |
| [班明江, 赵诺, 彭思利, 葛之葳, 邢玮, 李楠楠, 毛岭峰 (2024) 丛枝和外生菌根森林土壤微生物残体碳对有机碳贡献的整合分析. 生态学报, 44, 11254-11264.] | |
| [5] | Bardgett RD, Mommer L, De Vries FT (2014) Going underground: Root traits as drivers of ecosystem processes. Trends in Ecology & Evolution, 29, 692-699. |
| [6] |
Brown-Elliott BA, Wallace RJ Jr (2017) In vitro susceptibility testing of tedizolid against nontuberculous mycobacteria. Journal of Clinical Microbiology, 55, 1747-1754.
DOI PMID |
| [7] |
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annual Review of Plant Biology, 64, 807-838.
DOI PMID |
| [8] | Cai LJ, Guo ZH, Zhang JT, Gai ZJ, Liu JQ, Meng QY, Liu XH (2021) No tillage and residue mulching method on bacterial community diversity regulation in a black soil region of Northeastern China. PLoS ONE, 16, e0256970. |
| [9] | Chen ZH, Li YC, Chang SX, Xu QF, Li YF, Ma ZL, Qin H, Cai YJ (2021) Linking enhanced soil nitrogen mineralization to increased fungal decomposition capacity with Moso bamboo invasion of broadleaf forests. Science of the Total Environment, 771, 144779. |
| [10] | Cheng SL, Fang HJ, Xu M, Geng J, He S, Yu GX, Cao ZC (2018) Regulation of plant-soil-microbe interactions to soil organic carbon in natural ecosystems under elevated nitrogen deposition: A review. Acta Ecologica Sinica, 38, 8285-8295. (in Chinese with English abstract) |
| [程淑兰, 方华军, 徐梦, 耿静, 何舜, 于光夏, 曹子铖 (2018) 氮沉降增加情景下植物-土壤-微生物交互对自然生态系统土壤有机碳的调控研究进展. 生态学报, 38, 8285-8295.] | |
| [11] | Chu W, Guo XL, Zhang C, Zhou LT, Wu ZY, Lin WX (2022) Research progress and future directions of arbuscular mycorrhizal fungi-plant-rhizosphere microbial interaction. Chinese Journal of Eco-Agriculture, 30, 1709-1721. (in Chinese with English abstract) |
| [储薇, 郭信来, 张晨, 周柳婷, 吴则焰, 林文雄 (2022) 丛枝菌根真菌-植物-根际微生物互作研究进展与展望. 中国生态农业学报(中英文), 30, 1709-1721.] | |
| [12] | Ding H, Wang JH (2018) Study on the change trend of meteorological factors in Kuandian County from 1998 to 2017. Rural Technology, 30, 110-111. (in Chinese) |
| [丁桦, 王吉慧 (2018) 1998-2017年宽甸县气象因子变化趋势研究. 乡村科技, 30, 110-111.] | |
| [13] | Ding XJ, Jing RY, Huang YL, Chen BJ, Ma FY (2017) Bacterial structure and diversity of rhizosphere and bulk soil of Robinia pseudoacacia forests in Yellow River Delta. Acta Pedologica Sinica, 54, 1293-1302. (in Chinese with English abstract) |
| [丁新景, 敬如岩, 黄雅丽, 陈博杰, 马风云 (2017) 黄河三角洲刺槐根际与非根际细菌结构及多样性. 土壤学报, 54, 1293-1302.] | |
| [14] | Duan SL, Feng G, Limpens E, Bonfante P, Xie XN, Zhang L (2024) Cross-Kingdom nutrient exchange in the plant-arbuscular mycorrhizal fungus-bacterium continuum. Nature Reviews Microbiology, 22, 773-790. |
| [15] | Erktan A, Rillig MC, Carminati A, Jousset A, Scheu S (2020) Protists and collembolans alter microbial community composition, C dynamics and soil aggregation in simplified consumer-prey systems. Biogeosciences, 17, 4961-4980. |
| [16] | Esperschütz J, Zimmermann C, Dümig A, Welzl G, Buegger F, Elmer M, Munch JC, Schloter M (2013) Dynamics of microbial communities during decomposition of litter from pioneering plants in initial soil ecosystems. Biogeosciences, 10, 5115-5124. |
| [17] | Fontúrbel MT, Barreiro A, Vega JA, Martín A, Jiménez E, Carballas T, Fernández C, Díaz-Raviña M (2012) Effects of an experimental fire and post-fire stabilization treatments on soil microbial communities. Geoderma, 191, 51-60. |
| [18] | Fu LC, Ding ZJ, Tang M, Zeng H, Zhu B (2024) Rhizosphere effects of Betula platyphylla and Quercus mongolica and their seasonal dynamics in Dongling Mountain, Beijing. Chinese Journal of Plant Ecology, 48, 508-522. (in Chinese with English abstract) |
|
[付粱晨, 丁宗巨, 唐茂, 曾辉, 朱彪 (2024) 北京东灵山白桦和蒙古栎的根际效应及其季节动态. 植物生态学报, 48, 508-522.]
DOI |
|
| [19] | Ge W, Dong CB, Zhang ZY, Han YF, Liang ZQ (2021) Symbiotic interaction between ectomycorrhizal fungi and endobacteria: A review. Microbiology China, 48, 3810-3822. (in Chinese with English abstract) |
| [葛伟, 董醇波, 张芝元, 韩燕峰, 梁宗琦 (2021) 外生菌根真菌与内生细菌共生互作的研究进展. 微生物学通报, 48, 3810-3822.] | |
| [20] |
Ge Y, Xu SH, Xu Y (2019) Review on influencing factors of rhizosphere microbiome assemblage. Acta Agriculturae Zhejiangensis, 31, 2120-2130. (in Chinese with English abstract)
DOI |
|
[葛艺, 徐绍辉, 徐艳 (2019) 根际微生物组构建的影响因素研究进展. 浙江农业学报, 31, 2120-2130.]
DOI |
|
| [21] | Guo HB, Liu WY, Xie YQ, Wang ZY, Huang CT, Yi JF, Yang ZQ, Zhao JC, Yu XD, Sibirina LA (2024) Soil microbiome of shiro reveals the symbiotic relationship between Tricholoma bakamatsutake and Quercus mongolica. Frontiers in Microbiology, 15, 1361117. |
| [22] | He QH, Fang R, Zhang YQ, Xie YK, Tong XQ, Yang SZ (2022) The correlation of yield of various Stropharia rugosoannulata cultivated strains with forest environmental factors. Mycosystema, 41, 759-768. (in Chinese with English abstract) |
|
[何庆海, 方茹, 张燕琴, 谢宇凯, 童晓青, 杨少宗 (2022) 林下不同菌种皱环球盖菇产量与环境因子相关性分析. 菌物学报, 41, 759-768.]
DOI |
|
| [23] | Huang QY, Yang F, Xie LH, Cao HJ, Luo CY, Wang JF, Yan ZY, Ni HW (2021) Diversity and community structure of soil bacteria in different volcanoes, Wudalianchi. Acta Ecologica Sinica, 41, 8276-8284. (in Chinese with English abstract) |
| [黄庆阳, 杨帆, 谢立红, 曹宏杰, 罗春雨, 王继丰, 焉志远, 倪红伟 (2021) 五大连池火山土壤细菌多样性及其群落结构. 生态学报, 41, 8276-8284.] | |
| [24] | Huang SP, Jin DC, Li QL, Wei SL, Tang LH, Huang SM, Guo TX, Luo SM, Mo JY (2022) Study on the characteristics and common rule of bacteria community in banana rhizosphere and non-rhizosphere soil. Journal of Southern Agriculture, 53, 1253-1262. (in Chinese with English abstract) |
| [黄穗萍, 金德才, 李其利, 韦绍龙, 唐利华, 黄素梅, 郭堂勋, 罗述名, 莫贱友 (2022) 香蕉根际及非根际土壤细菌群落特征及共性规律研究. 南方农业学报, 53, 1253-1262.] | |
| [25] | Huang ZY, Su YG, Lin SN, Wu GP, Cheng H, Huang G (2023) Elevational patterns of microbial species richness and evenness across climatic zones and taxonomic scales. Ecology and Evolution, 13, e10594. |
| [26] | Huo XY, Ren CJ, Wang DX, Wu RQ, Wang YS, Li ZF, Huang DC, Qi HY (2023) Microbial community assembly and its influencing factors of secondary forests in Qinling Mountains. Soil Biology and Biochemistry, 184, 109075. |
| [27] | Ji RQ, Xing PJ, Xu Y, Li GL, Gao TT, Zhou JJ, Fu JH, Xie ML, Li Y (2019) Analyses on the composition of symbiotic fungi and bacteria in the roots and rhizosphere soil of Quercus mongolica. Mycosystema, 38, 1894-1906. (in Chinese with English abstract) |
|
[冀瑞卿, 邢鹏杰, 徐洋, 李冠霖, 高婷婷, 周吉江, 付俊慧, 谢孟乐, 李玉 (2019) 蒙古栎根内及根围土壤中共生真菌和细菌群落组成分析. 菌物学报, 38, 1894-1906.]
DOI |
|
| [28] |
Keiblinger KM, Hall EK, Wanek W, Szukics U, Hämmerle I, Ellersdorfer G, Böck S, Strauss J, Sterflinger K, Richter A, Zechmeister-Boltenstern S (2010) The effect of resource quantity and resource stoichiometry on microbial carbon-use-efficiency. FEMS Microbiology Ecology, 73, 430-440.
DOI PMID |
| [29] | Khan RAA, Najeeb S, Mao ZC, Ling J, Yang YH, Li Y, Xie BY (2020) Bioactive secondary metabolites from Trichoderma spp. against phytopathogenic bacteria and root-knot nematode. Microorganisms, 8, 401. |
| [30] |
Kundim BA, Itou Y, Sakagami Y, Fudou R, Iizuka T, Yamanaka S, Ojika M (2003) New haliangicin isomers, potent antifungal metabolites produced by a marine myxobacterium. Journal of Antibiotics, 56, 630-638.
DOI PMID |
| [31] | Kuzyakov Y (2010) Priming effects: Interactions between living and dead organic matter. Soil Biology and Biochemistry, 42, 1363-1371. |
| [32] | Li F, Ding HR, Yang YW, Liu YW, Wang H, Zhang Y, Li SP, Chen C, Tian DJ (2023) Field testing of pilot-scale bioslurry reactor for coking contaminated site: A case study in Anhui Province. Journal of Environmental Engineering Technology, 13, 1725-1731. (in Chinese with English abstract) |
| [李霏, 丁浩然, 杨乐巍, 刘渊文, 王恒, 张岳, 李书鹏, 陈成, 田德金 (2023) 安徽某焦化污染场地生物泥浆反应器中试研究案例. 环境工程技术学报, 13, 1725-1731.] | |
| [33] | Li FX, Huang YY, Wang ZJ, Shen JL, Sun J, Zhang YH, Wu X, Guo XN (2024) Corn cultivation alters bacterial diversity and function in salinized soils of the Diversionary Yellow River Irrigation Area. Chinese Journal of Eco-Agriculture, 32, 986-996. (in Chinese with English abstract) |
| [李凤霞, 黄业芸, 王长军, 沈靖丽, 孙娇, 张永宏, 吴霞, 郭鑫年 (2024) 玉米种植改变了引黄灌区盐渍化土壤细菌多样性与功能. 中国生态农业学报(中英文), 32, 986-996.] | |
| [34] | Li LL, Che ZH, Cao YH, Qi LL, Chen KL, Wang HS (2023) Analyzing the soil microbial characteristics of Poa alpigena Lindm. on bird island in Qinghai Lake based on metagenomics analysis. Water, 15, 239. |
| [35] | Li QX, Zhang S, Zhang FM, Ma YT, Wu L (2024) Research progress on the contribution of plant belowground carbon input to soil organic carbon based on bibliometrics. Chinese Journal of Soil Science, 55, 1777-1788. (in Chinese with English abstract) |
| [李清鑫, 张珊, 张复茂, 马燕天, 吴兰 (2024) 基于文献计量学的植物地下碳输入对土壤有机碳贡献的研究进展. 土壤通报, 55, 1777-1788.] | |
| [36] | Li XF, Han SJ, Guo ZL, Shao DK, Xin LH (2010) Changes in soil microbial biomass carbon and enzyme activities under elevated CO2 affect fine root decomposition processes in a Mongolian oak ecosystem. Soil Biology and Biochemistry, 42, 1101-1107. |
| [37] | Li XQ, Li Q, Duan YZ, Sun HQ, Chu H, Jia SB, Chen HJ, Tang WX (2024) Soil fungal communities varied across aspects of restored grassland in former mining areas of the Qinghai-Tibet Plateau. PLoS ONE, 19, e0295019. |
| [38] |
Ling N, Wang TT, Kuzyakov Y (2022) Rhizosphere bacteriome structure and functions. Nature Communications, 13, 836.
DOI PMID |
| [39] | Liu JW, Li XZ, Yao MJ (2021) Research progress on assembly of plant rhizosphere microbial community. Acta Microbiologica Sinica, 61, 231-248. (in Chinese with English abstract) |
| [刘京伟, 李香真, 姚敏杰 (2021) 植物根际微生物群落构建的研究进展. 微生物学报, 61, 231-248.] | |
| [40] | Liu LJ, Zhu QL, Wen DN, Yang L, Ni K, Xu XL, Cao JH, Meng L, Yang JL, Zhou JX, Zhu TB, Müller C (2024) Stimulation of organic N mineralization by N-acquiring enzyme activity alleviates soil microbial N limitation following afforestation in subtropical karst areas. Plant and Soil, 504, 879-894. |
| [41] |
Lu T, Xu NH, Lei CT, Zhang Q, Zhang ZY, Sun LW, He F, Zhou NY, Peñuelas J, Zhu YG, Qian HF (2023) Bacterial biogeography in China and its association to land use and soil organic carbon. Soil Ecology Letters, 5, 230172.
DOI |
| [42] | Luo CL, Hou L, Bai LL, Geng ZC, He WX (2017) Effects of forest thinning on soil CO2 emissions in a pine-oak mixed stand in the Qinling Mountains based on the Yasso 07 model estimate. Acta Ecologica Sinica, 37, 2894-2903. (in Chinese with English abstract) |
| [罗春林, 侯琳, 白龙龙, 耿增超, 和文祥 (2017) 抚育对林地土壤碳释放的影响——基于Yasso07估算. 生态学报, 37, 2894-2903.] | |
| [43] | Manici LM, Caputo F, Fornasier F, Paletto A, Ceotto E, De Meo I (2024) Ascomycota and Basidiomycota fungal Phyla as indicators of land use efficiency for soil organic carbon accrual with woody plantations. Ecological Indicators, 160, 111796. |
| [44] |
Nazir R, Warmink JA, Boersma H, van Elsas JD (2010) Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol Ecol, 71, 169-185.
DOI PMID |
| [45] |
Oshkin IY, Kulichevskaya IS, Rijpstra WIC, Damsté JSS, Rakitin AL, Ravin NV, Dedysh SN (2019) Granulicella sibirica sp. nov., a psychrotolerant acidobacterium isolated from an organic soil layer in forested tundra, West Siberia. International Journal of Systematic and Evolutionary Microbiology, 69, 1195-1201.
DOI PMID |
| [46] | Peng JG, Gong JY, Fan YH, Zhang H, Zhang YF, Bai YQ, Wang YM, Xie LJ (2022) Diversity of soil microbial communities in rhizosphere and non-rhizosphere of Rhododendron moulmainense. Scientia Silvae Sinicae, 58(2), 89-99. (in Chinese with English abstract) |
| [彭金根, 龚金玉, 范玉海, 张华, 张银凤, 白宇清, 王艳梅, 谢利娟 (2022) 毛棉杜鹃根际与非根际土壤微生物群落多样性. 林业科学, 58(2), 89-99.] | |
| [47] | Petrolli R, Vieira CA, Jakalski M, Bocayuva MF, Vallé C, Cruz ED, Selosse MA, Martos F, Kasuya MCM (2021) A fine-scale spatial analysis of fungal communities on tropical tree bark unveils the epiphytic rhizosphere in orchids. New Phytologist, 231, 2002-2014. |
| [48] | Puthusseri RM, Nair HP, Johny TK, Bhat SG (2021) Insights into the response of mangrove sediment microbiomes to heavy metal pollution: Ecological risk assessment and metagenomics perspectives. Journal of Environmental Management, 298, 113492. |
| [49] | Qin JF, Li Y, Liu GQ, Ai N, Liu CH (2022) Soil fungal community characteristics of seabuckthorn plantation with different restoration years in coal mine reclamation area. Chinese Journal of Soil Science, 53, 1413-1420. (in Chinese with English abstract) |
| [秦家凤, 李阳, 刘广全, 艾宁, 刘长海 (2022) 煤矿复垦区不同恢复年限沙棘人工林土壤真菌群落特征. 土壤通报, 53, 1413-1420.] | |
| [50] | Ran K, Wang ZJ, Li P (2023) Response difference of bacterial and fungal structure and diversity in rhizosphere soil of typical urban tree along urban-rural environmental gradient. Acta Ecologica Sinica, 43, 9758-9769. (in Chinese with English abstract) |
| [冉堃, 王泽锦, 李品 (2023) 典型城乡绿化树种土壤微生物组成和多样性的环境响应差异. 生态学报, 43, 9758-9769.] | |
| [51] | Reis IA, de Souza MG, Granja-Salcedo YT, Porcionato MAD, Prados LF, Siqueira GR, De Resende FD (2023) Effect of post-ruminal urea supply on growth performance of grazing nellore young bulls at dry season. Animals, 13, 207. |
| [52] | Ren H, Wang H, Qi X, Yu Z, Zheng X, Zhang S, Wang Z, Zhang M, Ahmed T, Li B (2021) The damage caused by decline disease in bayberry plants through changes in soil properties, rhizosphere microbial community structure and metabolites. Plants, 10, 2083. |
| [53] | Rezaei F, Joh LD, Berry AM, VanderGheynst JS (2011) Xylanase and cellulase production by Acidothermus cellulolyticus grown on switchgrass in solid-state fermentation. Biofuels, 2, 21-32. |
| [54] | Sergentani AG, Gonou-Zagou Z, Kapsanaki-Gotsi E, Hatzinikolaou DG (2016) Lignocellulose degradation potential of Basidiomycota from Thrace (NE Greece). International Biodeterioration & Biodegradation, 114, 268-277. |
| [55] | Sheng YZ, Huang L, Ye PS, Lai J, Liu Y, Zhang QF, Liu J, Li FS, Wei SG (2024) Rhizosphere and non-rhizosphere soil microbial community structure and diversity Asparagus officinalis L. Journal of Sichuan Agricultural University, 42, 330-338. (in Chinese with English abstract) |
| [盛玉珍, 黄玲, 叶鹏盛, 赖佳, 刘勇, 张骞方, 刘佳, 李丰山, 韦树谷 (2024) 芦笋根际与非根际土壤微生物群落结构及多样性特征. 四川农业大学学报, 42, 330-338.] | |
| [56] | Shi LL, Fu SL (2014) Review of soil biodiversity research: History, current status and future challenges. Chinese Science Bulletin, 59, 493-509. (in Chinese with English abstract) |
| [时雷雷, 傅声雷 (2014) 土壤生物多样性研究:历史、现状与挑战. 科学通报, 59, 493-509.] | |
| [57] | Si YJ, Xu Y, Li BQ, Liu J, Meng LP, Li Y, Ji RQ, Liu SY (2022) Ectomycorrhizospheric microbiome assembly rules of Quercus mongolica in the habitat of Songrong (Tricholoma matsutake) and the effect of neighboring plants. Diversity, 14, 810. |
| [58] | Soares M, Rousk J (2019) Microbial growth and carbon use efficiency in soil: Links to fungal-bacterial dominance, SOC-quality and stoichiometry. Soil Biology and Biochemistry, 131, 195-205. |
| [59] | Sokol N, Foley M, Blazewicz SJ, Battacharyya A, Estera-Molina K, Firestone MK, Greenlon A, Hungate BA, Kimbrel J, Liquet J, Lafler M, Marple M, Nico P, Slessarev E, Pett-Ridge J (2022) Divergent microbial traits influence the transformation of living versus dead root inputs to soil carbon. BioRxiv, doi: 10.1101/2022.09.02.506384. |
| [60] | Soop K, Wallace M, Dima B (2018) New Cortinarius (Agaricales) species described from New Zealand. New Zealand Journal of Botany, 56, 163-182. |
| [61] | Tong T, Ji RT, Xu QJ, Wang JG, Li XO, Zhang Y (2024) Effects of activated sludge extraction on rhizosphere soil microbial community structure of rice. Journal of Environmental Engineering Technology, 14, 148-157. (in Chinese with English abstract) |
| [童彤, 纪荣婷, 许秋瑾, 王建国, 李小鸥, 张悦 (2024) 活性污泥萃取液施用对水稻根际土壤微生物群落结构的影响. 环境工程技术学报, 14, 148-157.] | |
| [62] | Wang CQ, Kuzyakov Y (2024) Mechanisms and implications of bacterial-fungal competition for soil resources. ISME Journal, 18, wrae073. |
| [63] | Wang H, Xu KP, Tian GS (2018) Review on genus Lactarius. Central South Pharmacy, 16, 504-512. (in Chinese with English abstract) |
| [王和, 徐康平, 谭桂山 (2018) 乳菇属真菌研究概况. 中南药学, 16, 504-512.] | |
| [64] | Wang QT, Zhu WR, Liu ML, Wang HT, Wang YP, Zhang GC, Jiang YZ (2015) Comparison on bacterial community of rhizosphere and bulk soil of poplar plantation based on pyrosequencing. Chinese Journal of Applied and Environmental Biology, 21, 967-973. (in Chinese with English abstract) |
| [汪其同, 朱婉芮, 刘梦玲, 王华田, 王延平, 张光灿, 姜岳忠 (2015) 基于高通量测序的杨树人工林根际和非根际细菌群落结构比较. 应用与环境生物学报, 21, 967-973.] | |
| [65] | Wang XL, Yao B, Su XY (2018) Linking enzymatic oxidative degradation of lignin to organics detoxification. International Journal of Molecular Sciences, 19, 3373. |
| [66] | Wang YL, Wang R, Lu B, Guerin-Laguette A, He XH, Yu FQ (2021) Mycorrhization of Quercus mongolica seedlings by Tuber melanosporum alters root carbon exudation and rhizosphere bacterial communities. Plant and Soil, 467, 391-403. |
| [67] | Wei SL, Fang J, Zhang TJ, Wang JG, Cheng YC, Ma J, Xie R, Liu ZX, Su EH, Ren YF, Zhao XQ, Zhang XQ, Lu ZY (2023) Dynamic changes of soil microorganisms in rotation farmland at the western foot of the Greater Khingan Range. Frontiers in Bioengineering and Biotechnology, 11, 1191240. |
| [68] | Wei YY, Cui LJ, Zhang MY, Pan X (2019) Research advances in microbial mechanisms underlying priming effect of soil organic carbon mineralization. Chinese Journal of Ecology, 38, 1202-1211. (in Chinese with English abstract) |
| [魏圆云, 崔丽娟, 张曼胤, 潘旭 (2019) 土壤有机碳矿化激发效应的微生物机制研究进展. 生态学杂志, 38, 1202-1211.] | |
| [69] | Yin D, Zhu YW, Hu M, Xu L, Yu HY (2024) Rice rhizosphere microbiomes and their driving cycling of soil carbon, nitrogen, and phosphorus. Journal of Plant Nutrition and Fertilizers, 30, 2207-2220. (in Chinese with English abstract) |
| [尹丹, 朱忆雯, 胡敏, 徐乐, 于焕云 (2024) 水稻根际微生物及其驱动的土壤碳氮磷循环. 植物营养与肥料学报, 30, 2207-2220.] | |
| [70] | Yu HY, Zheng J, Lu QJ, Zhang YT, Yu HO, Han Y (2023) Present situation and management suggestions of Quercus mongolica forest resources in Jiaohe City. Journal of Jilin Forestry Science and Technology, 52(4), 39-42. (in Chinese with English abstract) |
| [于海媛, 郑军, 卢庆杰, 张义涛, 于海鸥, 韩勇 (2023) 蛟河市蒙古栎林资源现状分析与经营建议. 吉林林业科技, 52(4), 39-42.] | |
| [71] | Zeng Q, Liu SY, Xiang JF, Liang ZH, Zhai CC, Yao BM, Han LL, Ge AH, Zhang LM (2024) Microbial communities in the rhizosphere of watermelon varieties resistant and susceptible to Fusarium wilt: Differences and relationship with disease occurrence. Acta Microbiologica Sinica, 64, 2882-2900. (in Chinese with English abstract) |
| [曾青, 刘四义, 向吉方, 梁志怀, 翟常春, 姚保民, 韩丽丽, 葛安辉, 张丽梅 (2024) 西瓜枯萎病抗感品种根际微生物群落特征差异及其与病害发生的关系. 微生物学报, 64, 2882-2900.] | |
| [72] |
Zeng Q, Xiong C, Yin M, Ge AH, Han LL, Zhang LM (2023) Research progress on ecological functions and community assembly of plant microbiomes. Biodiversity Science, 31, 22667. (in Chinese with English abstract)
DOI |
|
[曾青, 熊超, 尹梅, 葛安辉, 韩丽丽, 张丽梅 (2023) 植物微生物组生态功能与群落构建过程研究进展. 生物多样性, 31, 22667.]
DOI |
|
| [73] | Zhang EP, Tian YY, Li M, Shi M, Jiang YH, Ren RB, Zhang SH (2018) Effects of various long-term fertilization regimes on soil microbial functional diversity in tomato rhizosphere soil. Acta Ecologica Sinica, 38, 5027-5036. (in Chinese with English abstract) |
| [张恩平, 田悦悦, 李猛, 时毛, 蒋雨含, 任如冰, 张淑红 (2018) 长期不同施肥对番茄根际土壤微生物功能多样性的影响. 生态学报, 38, 5027-5036.] | |
| [74] |
Zhang J, Kong HZ, Huang XL, Fu SL, Guo LD, Guo QH, Lei FM, Lü Z, Zhou YR, Ma KP (2022) Thirty key questions for biodiversity science in China. Biodiversity Science, 30, 22609. (in Chinese with English abstract)
DOI |
|
[张健, 孔宏智, 黄晓磊, 傅声雷, 郭良栋, 郭庆华, 雷富民, 吕植, 周玉荣, 马克平 (2022) 中国生物多样性研究的30个核心问题. 生物多样性, 30, 22609.]
DOI |
|
| [75] | Zhao Y, Hao B, Xu H, Zhang YZ, Wang YZ, Li XY, Yao ZY, Li JJ, Zhang CT (2019) Fauna resource investigation of Tachinidae (Diptera) from Mt. Huangyi, eastern Liaoning, China. Journal of Environmental Entomology, 41, 1208-1217. (in Chinese with English abstract) |
| [赵颍, 郝博, 徐浩, 张怡卓, 王玉卓, 李辛夷, 姚志远, 李君健, 张春田 (2019) 辽宁东部黄椅山寄蝇科昆虫资源调查. 环境昆虫学报, 41, 1208-1217.] | |
| [76] | Zhu P, Liu WY, Liu ZH, Jiang BH, Xu JQ, Qu YL, Sun ZY, Bai XF, Hou YP (2024) Community structure and diversity of soil bacteria in different common forest types in Kunyu Mountain. Acta Scientiarum Naturalium Universitatis Sunyatseni, 63(6), 132-140. (in Chinese with English abstract) |
| [朱萍, 刘文燕, 刘展航, 蒋博涵, 许嘉庆, 曲彦霖, 孙中元, 柏新富, 侯玉平 (2024) 昆嵛山常见林型土壤细菌的群落结构及多样性分析. 中山大学学报(自然科学版中英文), 63(6), 132-140.] | |
| [77] | Zhu Y, Wang YX, Qin FC, Meng S, Wang SK, Lu JK (2024) Composition and function of soil microbial community in rhizosphere soil and bulk soil of Eucalyptus plantation across different stand ages. Aeta Eeologica Siniea, 44, 8409-8422. (in Chinese with English abstract) |
| [朱媛, 王亚鑫, 覃方锉, 孟森, 王胜坤, 陆俊锟 (2024) 不同林龄桉树根际及非根际土壤微生物群落结构及功能. 生态学报, 44, 8409-8422.] |
| [1] | Guohong Liu, Bo Liu, Jianmei Che, Qianqian Chen, Naiquan Lin, Weidong Cui. Diversity of Bacillus-like species isolated from potato rhizosphere soils in Yili, Xinjiang [J]. Biodiv Sci, 2017, 25(8): 856-863. |
| [2] | Chunnan Li, Hairui Cui, Weibo Wang. Genetic diversity in rhizosphere soil microbes detected with SRAP markers [J]. Biodiv Sci, 2011, 19(4): 485-493. |
| [3] | Jie Zhang, Di Wu, Chunlei Wang, Hongjun Qu, Xuezhong Zou, Chuanping Yang. Genetic diversity analysis of Quercus mongolica populations with In-ter-Simple Sequence Repeats (ISSR) technique [J]. Biodiv Sci, 2007, 15(3): 292-299. |
| [4] | Lixia Zhou, Mingmao Ding. Soil microbial characteristics as bioindicators of soil health [J]. Biodiv Sci, 2007, 15(2): 162-171. |
| [5] | YU Shun-Li, LIU Can-Ran, MA Ke-Ping. A study on the ecotones betweenQuercus mongolica community and other communities [J]. Biodiv Sci, 2000, 08(3): 277-283. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()