



Biodiv Sci ›› 2010, Vol. 18 ›› Issue (3): 233-240. DOI: 10.3724/SP.J.1003.2010.233 cstr: 32101.14.SP.J.1003.2010.233
• Special Issue • Previous Articles Next Articles
Junhong Zhang1, Huahong Huang1, Zaikang Tong1,*( ), Longjun Cheng1, Yuelong Liang2, Yiliang Chen3
), Longjun Cheng1, Yuelong Liang2, Yiliang Chen3
Received:2009-11-11
Accepted:2010-04-24
Online:2010-05-20
Published:2012-02-08
Contact:
Zaikang Tong
Junhong Zhang, Huahong Huang, Zaikang Tong, Longjun Cheng, Yuelong Liang, Yiliang Chen. Genetic diversity in six natural populations of Betula luminifera from southern China[J]. Biodiv Sci, 2010, 18(3): 233-240.
| 群体 Population | 采样地 Collection site | 经纬度 Locality | 温度 Temperature (℃) | 降水量 Precipitation (mm) |
|---|---|---|---|---|
| 浙江临安 LA | 浙江临安太湖源 Taihuyuan, Lin’an, Zhejiang | 119°33′ E 30°25′ N | 15.5 | 1,375.8 |
| 浙江庆元 QY | 浙江庆元巾子峰 Jinzifeng, Qingyuan, Zhejiang | 119°08′ E 27°41′ N | 17.7 | 1,645.5 |
| 江西龙南 LN | 江西赣州九连山 Jiulianshan, Ganzhou, Jiangxi | 114°28′ E 24°33′ N | 19.5 | 1,395.3 |
| 广西龙胜 LS | 广西龙胜花坪 Huaping, Longsheng, Guangxi | 109°55′ E 25°38′ N | 18.7 | 1,577.3 |
| 贵州修文 XW | 贵州修文 Xiuwen, Guizhou | 106°34′ E 26°49′ N | 15.2 | 1,128.3 |
| 福建尤溪 YX | 福建尤溪 Youxi, Fujian | 118°10′ E 25°56′ N | 19.3 | 1,322.8 |
Table 1 Survey of sampling locations in natural populations of Betula luminifera
| 群体 Population | 采样地 Collection site | 经纬度 Locality | 温度 Temperature (℃) | 降水量 Precipitation (mm) |
|---|---|---|---|---|
| 浙江临安 LA | 浙江临安太湖源 Taihuyuan, Lin’an, Zhejiang | 119°33′ E 30°25′ N | 15.5 | 1,375.8 |
| 浙江庆元 QY | 浙江庆元巾子峰 Jinzifeng, Qingyuan, Zhejiang | 119°08′ E 27°41′ N | 17.7 | 1,645.5 |
| 江西龙南 LN | 江西赣州九连山 Jiulianshan, Ganzhou, Jiangxi | 114°28′ E 24°33′ N | 19.5 | 1,395.3 |
| 广西龙胜 LS | 广西龙胜花坪 Huaping, Longsheng, Guangxi | 109°55′ E 25°38′ N | 18.7 | 1,577.3 |
| 贵州修文 XW | 贵州修文 Xiuwen, Guizhou | 106°34′ E 26°49′ N | 15.2 | 1,128.3 |
| 福建尤溪 YX | 福建尤溪 Youxi, Fujian | 118°10′ E 25°56′ N | 19.3 | 1,322.8 |
| 群体 Population | 样本数量 Sample size | 多态位点数 N | 多态位点百分率 PPL(%) | Nei’s基因多样性(标准差) hj(standard error) | 个体取样的变异比例 VarI (%) | 位点导致的变异比例 VarL (%) |
|---|---|---|---|---|---|---|
| 浙江临安 LA | 20 | 337 | 94.9 | 0.3422(0.0074) | 40.3 | 59.7 |
| 浙江庆元 QY | 20 | 350 | 98.6 | 0.3645(0.0065) | 46.5 | 53.5 |
| 江西龙南 LN | 20 | 333 | 93.8 | 0.3309(0.0072) | 37.5 | 62.5 |
| 广西龙胜 LS | 20 | 332 | 93.5 | 0.3143(0.0076) | 37.6 | 62.4 |
| 贵州修文 XW | 20 | 331 | 93.2 | 0.3230(0.0076) | 35.1 | 64.9 |
| 福建尤溪 YX | 20 | 335 | 94.4 | 0.3535(0.0069) | 37.7 | 62.3 |
Table 2 Genetic diversity of six Betula luminifera populations detected by AFLP
| 群体 Population | 样本数量 Sample size | 多态位点数 N | 多态位点百分率 PPL(%) | Nei’s基因多样性(标准差) hj(standard error) | 个体取样的变异比例 VarI (%) | 位点导致的变异比例 VarL (%) |
|---|---|---|---|---|---|---|
| 浙江临安 LA | 20 | 337 | 94.9 | 0.3422(0.0074) | 40.3 | 59.7 |
| 浙江庆元 QY | 20 | 350 | 98.6 | 0.3645(0.0065) | 46.5 | 53.5 |
| 江西龙南 LN | 20 | 333 | 93.8 | 0.3309(0.0072) | 37.5 | 62.5 |
| 广西龙胜 LS | 20 | 332 | 93.5 | 0.3143(0.0076) | 37.6 | 62.4 |
| 贵州修文 XW | 20 | 331 | 93.2 | 0.3230(0.0076) | 35.1 | 64.9 |
| 福建尤溪 YX | 20 | 335 | 94.4 | 0.3535(0.0069) | 37.7 | 62.3 |
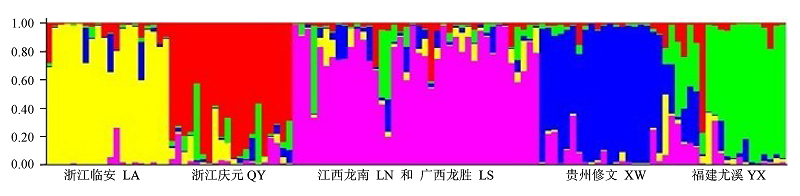
Fig. 1 Ancestry of Betula luminifera from six population estimated using STRUCTURE. Population LA, QY, admixture of LN and LS, XW and YX gene pools are shown in yellow, red, pink, blue and green respectively. Y-coordinate denotes the proportion of ancestry components in an individual in relation to other population.
| 变异来源 Source of variance | 自由度 df | 变异组分 Variance components | 变异贡献率(%) Contribution of variation | 遗传分化系数(фST) Genetic differentiation index | P |
|---|---|---|---|---|---|
| 群体间 Among populations | 5 | 6.38404 | 11.49% | 0.11489 | <0.0001** |
| 群体内 Within populations | 113 | 49.18305 | 88.51% | ||
| 总计 Total | 118 | 55.56709 |
Table 3 Analysis of molecular variance (AMOVA) of six populations in Betula luminifera
| 变异来源 Source of variance | 自由度 df | 变异组分 Variance components | 变异贡献率(%) Contribution of variation | 遗传分化系数(фST) Genetic differentiation index | P |
|---|---|---|---|---|---|
| 群体间 Among populations | 5 | 6.38404 | 11.49% | 0.11489 | <0.0001** |
| 群体内 Within populations | 113 | 49.18305 | 88.51% | ||
| 总计 Total | 118 | 55.56709 |
| 群体 | LA | QY | LN | LS | XW | YX |
|---|---|---|---|---|---|---|
| LA | **** | 0.0637 | 0.1115 | 0.1135 | 0.1146 | 0.0881 |
| QY | 0.0377 | **** | 0.0534 | 0.0516 | 0.0612 | 0.0358 |
| LN | 0.0657 | 0.0302 | **** | 0.0349 | 0.0513 | 0.0385 |
| LS | 0.0643 | 0.0276 | 0.0173 | **** | 0.0551 | 0.0552 |
| XW | 0.0665 | 0.0342 | 0.0266 | 0.0276 | **** | 0.0379 |
| YX | 0.0529 | 0.0210 | 0.0209 | 0.0293 | 0.0201 | **** |
Table 4 Nei’s genetic distance (below diagonal) and genetic differentiation (pairwise фST,above diagonal) between populations in Betula luminifera
| 群体 | LA | QY | LN | LS | XW | YX |
|---|---|---|---|---|---|---|
| LA | **** | 0.0637 | 0.1115 | 0.1135 | 0.1146 | 0.0881 |
| QY | 0.0377 | **** | 0.0534 | 0.0516 | 0.0612 | 0.0358 |
| LN | 0.0657 | 0.0302 | **** | 0.0349 | 0.0513 | 0.0385 |
| LS | 0.0643 | 0.0276 | 0.0173 | **** | 0.0551 | 0.0552 |
| XW | 0.0665 | 0.0342 | 0.0266 | 0.0276 | **** | 0.0379 |
| YX | 0.0529 | 0.0210 | 0.0209 | 0.0293 | 0.0201 | **** |
| [1] | Chen W (陈伟) (2006) Genetic diversity of the natural populations of Betula luminifera. Journal of Beijing Forestry University (北京林业大学学报), 28 (6),28-34. (in Chinese with English abstract) |
| [2] | Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus, 12, 13-14. |
| [3] |
Evanno G, Regnaut S, Goudetd J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611-2620.
DOI URL PMID |
| [4] | Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics (Online), 1, 47-50. |
| [5] |
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics, 164, 1567-1587.
URL PMID |
| [6] | Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes, 7, 574-578. |
| [7] | Feng FJ, Han SJ, Wang HM (2006) Genetic diversity and genetic differentiation of natural Pinus koraiensis population. Journal of Forestry Research, 17, 21-24. |
| [8] | Gao YK (高亦珂), Nie SQ (聂绍荃), Zu YG (祖元刚) (1999) Genetic structure analysis by RAPD in Betula platyphylla nature population in Northeast of China. In: The Application, Method and Theory of Molecular Ecology (分子生态学理论、方法和应用), pp.196-205. China Higher Education Press, Beijing. (in Chinese) |
| [9] |
Gillies ACM, Navarro C, Lowe AJ, Newton AC, Hernandez M, Wilson J, Cornelius JP (1999) Genetic diversity in Mesoamerican populations of mahogany (Swietenia macrophylla), assessed using RAPDs. Heredity, 83, 722-732.
URL PMID |
| [10] | Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Plant Population Genetics, Breeding and Genetic Resources (eds Brown AHD, Clegg MT, Kahler AL,Weir BS), pp.43-63. Sinauer Associates, Sunderland. |
| [11] | Hamrick JL, Godt MJW, Sherman-Broyes SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forests, 6, 95-124. |
| [12] |
He JS, Chen L, Si Y, Huang B, Ban XQ, Wang YW (2008) Population structure and genetic diversity distribution in wild and cultivated populations of the traditional Chinese medicinal plant Magnolia officinalis subsp. biloba (Magnoliaceae). Genetica, 135, 233-243.
URL PMID |
| [13] | Li QM (李巧明), Zhao JL (赵建立) (2007) Genetic diversity of Phyllanthus embilica populations in dry-hot valleys in Yunnan. Biodiversity Science (生物多样性), 15, 84-91. (in Chinese with English abstract) |
| [14] | Luo JX (罗建勋), Gu WC (顾万春), Chen SY (陈少瑜) (2006) Allozyme variation in 10 natural population of Picea asperata. Acta Phytoecologica Sinica (植物生态学报), 30, 165-173. (in Chinese with English abstract) |
| [15] | Miller MP (1997) Tools for population genetic analyses (TFPGA) version 1.3: A windows program for the analysis of allozyme and molecular genetic data. Department of Biological Sciences, Northern Arizona University, Flagstaff. |
| [16] | Nei M (1972) Genetic distance between populations. The American Naturalist, 106, 283-292. |
| [17] | Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583-590. |
| [18] | Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, USA, 76, 5269-5273. |
| [19] | Powell W, Thomas WTB, Baird E (1997) Analysis of quantitative traits in barley by the use of amplified fragment polymorphism. Heredity, 79, 48-59. |
| [20] |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945-959.
URL PMID |
| [21] | de la Torre A, López C, Yglesias E, Cornelius JP (2008) Genetic (AFLP) diversity of nine Cedrela odorata populations in Madre de Dios, southern Peruvian Amazon. Forest Ecology and Management, 255, 334-339. |
| [22] |
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology, 11, 139-151.
DOI URL PMID |
| [23] | Wang ZF (王峥峰), Ge XJ (葛学军) (2009) Not only genetic diversity: advances in plant conservation genetics. Biodiversity Science (生物多样性), 17, 330-339. (in Chinese with English abstract) |
| [24] | Wu ZC (吴子诚), Wang LH (王乐辉) (1996) The characteristics of selective population and improved technology of Betula luminifera. Journal of Sichuan Forestry Science and Technology (四川林业科技), 17(4),17-28. (in Chinese with English abstract) |
| [25] | Xie YQ (谢一青), Li ZZ (李志真), Huang RZ (黄儒珠), Xiao XX (肖祥希), Huang Y (黄勇) (2008) Genetic diversity of Betula luminifera populations at different altitudes in Wuyi Mountains and its association with ecological factors. Scientia Silvae Sinicae (林业科学), 44(3),50-55. (in Chinese with English abstract) |
| [26] | Yeh FC, Chong DKX, Yang RC (1995) RAPD variation within and among natural populations of trembling aspen ( Populus tremuloides) from Alberta. Heredity, 86, 454-460. |
| [27] | You WY (尤卫艳), Huang HH (黄华宏), Tong ZK (童再康), Zhu YQ (朱玉球) (2008) Establishment of AFLP molecular labeling technique system for Betula luminifera. Biotechnology (生物技术), 18(6),42-47. (in Chinese with English abstract) |
| [28] | Zabeau M, Vos P (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application, EP 534858A1. |
| [29] | Zeng J (曾杰), Wang ZR (王中仁), Zhou SL (周世良), Zheng HS (郑海水), Bai JY (白嘉雨) (2003) Allozymed diversity in natural populations of Betula alnoides from Guangxi, China. Acta Phytoecologica Sinica (植物生态学报), 27, 66-72. (in Chinese with English abstract) |
| [30] | Zeng J, Zou YP, Bai JY, Zheng HS (2003) RAPD analysis of genetic variation in natural populations of Betula alnoides from Guangxi, China. Euphytica, 134, 33-41. |
| [31] | Zhang P (张萍), Zhou ZC (周志春), Jin GQ (金国庆), Fan HH (范辉华), Hu HB (胡红宝) (2006) Genetic diversity analysis and provenance zone allocation of Schima superba in China using RAPD markers. Scientia Silvae Sinicae (林业科学), 42(2),38-42. (in Chinese with English abstract) |
| [32] | Zheng WJ (郑万钧) (1985) Records of Chinese Trees (Vol.2)(中国树木志)(第二卷), pp.2124-2131. China Forestry Publishing House, Beijing. (in Chinese) |
| [33] | Zheng J (郑健), Zheng YQ (郑勇奇), Zhang CH (张川红), Zong YC (宗亦臣), Li BJ (李伯菁), Wu C (吴超) (2008) Genetic diversity in natural populations of Sorbus pohuashanensis. Biodiversity Science (生物多样性), 16, 562-569. (in Chinese with English abstract) |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Hong Deng, Zhanyou Zhong, Chunni Kou, Shuli Zhu, Yuefei Li, Yuguo Xia, Zhi Wu, Jie Li, Weitao Chen. Population genetic structure and evolutionary history of Hemibagrus guttatus based on mitochondrial genomes [J]. Biodiv Sci, 2025, 33(1): 24241-. |
| [3] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [4] | Xianglin Yang, Caiyun Zhao, Junsheng Li, Fangfang Chong, Wenjin Li. Invasive plant species lead to a more clustered community phylogenetic structure: An analysis of herbaceous plants in Guangxi’s national nature reserves [J]. Biodiv Sci, 2024, 32(11): 24175-. |
| [5] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [6] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [7] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [8] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [9] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [10] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [11] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [12] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [13] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [14] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [15] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()