



生物多样性 ›› 2010, Vol. 18 ›› Issue (3): 251-261. DOI: 10.3724/SP.J.1003.2010.251 cstr: 32101.14.SP.J.1003.2010.251
赵亮1,2, 张洁2, 刘志瑾2, 许木启2, 李明2,*( )
)
收稿日期:2009-12-28
接受日期:2010-05-11
出版日期:2010-05-20
发布日期:2012-02-08
通讯作者:
李明
作者简介: E-mail: lim@ioz.ac.cn基金资助:
Liang Zhao1,2, Jie Zhang2, Zhijin Liu2, Muqi Xu2, Ming Li2,*( )
)
Received:2009-12-28
Accepted:2010-05-11
Online:2010-05-20
Published:2012-02-08
Contact:
Ming Li
摘要:
为促进乔氏新银鱼(Neosalanx jordani)种质资源的保护, 采集了长江流域和淮河流域5个乔氏新银鱼地理种群计129个样本, 利用线粒体细胞色素b基因(Cyt b)全序列作为分子标记, 初步分析了乔氏新银鱼种群的遗传多样性、遗传结构及种群历史动态。研究结果共检测到18个Cyt b单倍型, 发现和其他鱼类相比, 乔氏新银鱼具有较高的单倍型多样性(h, 0.590±0.047), 但核苷酸多样性较低(π, 0.00088±0.00011)。分子变异分析(AMOVA)表明, 乔氏新银鱼5个地理种群内个体间和流域内种群间均存在显著的遗传差异, 而长江流域与淮河流域之间遗传分化不明显, 显示乔氏新银鱼遗传分化与当前水系的分布格局不吻合。结果表明乔氏新银鱼目前的遗传格局主要是由于长距离独立的建群事件、基因流限制以及种群的持续扩张的共同作用而形成的。种群历史动态分析结果显示乔氏新银鱼种群为近期扩张种群, 其大部分变异发生在1.897万年之内, 与最后一次冰期全面消退、海平面上升、长江流域及淮河流域中下游大量湖泊(适宜生境)形成的时间相吻合。建议对现存的乔氏新银鱼种群, 特别是遗传多样性较高的那些种群(如鄱阳湖、太湖及洪泽湖种群)分别保护。
赵亮, 张洁, 刘志瑾, 许木启, 李明 (2010) 乔氏新银鱼基于细胞色素b序列的种群遗传结构和种群历史. 生物多样性, 18, 251-261. DOI: 10.3724/SP.J.1003.2010.251.
Liang Zhao, Jie Zhang, Zhijin Liu, Muqi Xu, Ming Li (2010) Population genetic structure and demographic history of Neosalanx jordanibased on cytochrome b sequences. Biodiversity Science, 18, 251-261. DOI: 10.3724/SP.J.1003.2010.251.
| 单倍型 Haplotypes | 长江流域 Yangtze River Basin | 淮河流域 Huaihe River Basin | 合计 Total | ||||
|---|---|---|---|---|---|---|---|
| 太湖 Taihu lake | 南漪湖 Nanyihu lake | 鄱阳湖 Poyanghu lake | 洪泽湖 Hongzehu lake | 瓦埠湖 Wabuhu lake | |||
| M01 | 1 | 1 | |||||
| M02 | 1 | 1 | |||||
| M03 | 3 | 3 | |||||
| M04 | 2 | 2 | |||||
| M05 | 1 | 1 | |||||
| M06 | 1 | 1 | |||||
| M07 | 1 | 1 | |||||
| M08 | 1 | 1 | |||||
| M09 | 4 | 4 | |||||
| M10 | 1 | 1 | |||||
| M11 | 1 | 1 | |||||
| M12 | 4 | 15 | 2 | 21 | |||
| M13 | 3 | 3 | |||||
| M14 | 2 | 2 | |||||
| M15 | 1 | 1 | |||||
| M16 | 1 | 1 | 2 | ||||
| M17 | 17 | 23 | 3 | 16 | 21 | 80 | |
| M18 | 3 | 3 | |||||
| 样本量 Sample number | 30 | 25 | 24 | 25 | 25 | 129 | |
表1 线粒体细胞色素b单倍型在乔氏新银鱼各种群中的分布及数量
Table 1 Distribution and the number of mtDNA Cyt b haplotypes in each population ofNeosalanx jordani
| 单倍型 Haplotypes | 长江流域 Yangtze River Basin | 淮河流域 Huaihe River Basin | 合计 Total | ||||
|---|---|---|---|---|---|---|---|
| 太湖 Taihu lake | 南漪湖 Nanyihu lake | 鄱阳湖 Poyanghu lake | 洪泽湖 Hongzehu lake | 瓦埠湖 Wabuhu lake | |||
| M01 | 1 | 1 | |||||
| M02 | 1 | 1 | |||||
| M03 | 3 | 3 | |||||
| M04 | 2 | 2 | |||||
| M05 | 1 | 1 | |||||
| M06 | 1 | 1 | |||||
| M07 | 1 | 1 | |||||
| M08 | 1 | 1 | |||||
| M09 | 4 | 4 | |||||
| M10 | 1 | 1 | |||||
| M11 | 1 | 1 | |||||
| M12 | 4 | 15 | 2 | 21 | |||
| M13 | 3 | 3 | |||||
| M14 | 2 | 2 | |||||
| M15 | 1 | 1 | |||||
| M16 | 1 | 1 | 2 | ||||
| M17 | 17 | 23 | 3 | 16 | 21 | 80 | |
| M18 | 3 | 3 | |||||
| 样本量 Sample number | 30 | 25 | 24 | 25 | 25 | 129 | |
| 种群 Population | 样本量 Number of samples (N) | 单倍型数量 Number of haplotypes (H) | 单倍型多样性 Haplotype diversity (h±SD) | 多态性位点数 Number of polymorphic sites (S) | 核苷酸多样性 Nucleotide diversity (π±SD) |
|---|---|---|---|---|---|
| 长江流域 Yangtze River Basin | |||||
| 太湖 Taihu lake | 30 | 8 | 0.664±0.089 | 7 | 0.00077±0.00015 |
| 南漪湖 Nanyihu lake | 25 | 3 | 0.157±0.096 | 5 | 0.00035±0.00022 |
| 鄱阳湖 Poyanghu lake | 24 | 5 | 0.587±0.102 | 5 | 0.00085±0.00019 |
| 小计 Subtotal | 79 | 13 | 0.648±0.049 | 16 | 0.00093±0.00013 |
| 淮河流域 Huaihe River Basin | |||||
| 洪泽湖 Hongzehu lake | 25 | 6 | 0.583±0.109 | 6 | 0.00067±0.00017 |
| 瓦埠湖 Wabuhu lake | 25 | 3 | 0.290±0.109 | 5 | 0.00072±0.00028 |
| 小计 Subtotal | 50 | 7 | 0.449±0.086 | 9 | 0.00071±0.00018 |
| 总体 All samples | 129 | 18 | 0.590±0.047 | 24 | 0.00088±0.00011 |
表2 乔氏新银鱼种群遗传多样性
Table 2 Genetic diversity indices of Neosalanx jordanipopulations
| 种群 Population | 样本量 Number of samples (N) | 单倍型数量 Number of haplotypes (H) | 单倍型多样性 Haplotype diversity (h±SD) | 多态性位点数 Number of polymorphic sites (S) | 核苷酸多样性 Nucleotide diversity (π±SD) |
|---|---|---|---|---|---|
| 长江流域 Yangtze River Basin | |||||
| 太湖 Taihu lake | 30 | 8 | 0.664±0.089 | 7 | 0.00077±0.00015 |
| 南漪湖 Nanyihu lake | 25 | 3 | 0.157±0.096 | 5 | 0.00035±0.00022 |
| 鄱阳湖 Poyanghu lake | 24 | 5 | 0.587±0.102 | 5 | 0.00085±0.00019 |
| 小计 Subtotal | 79 | 13 | 0.648±0.049 | 16 | 0.00093±0.00013 |
| 淮河流域 Huaihe River Basin | |||||
| 洪泽湖 Hongzehu lake | 25 | 6 | 0.583±0.109 | 6 | 0.00067±0.00017 |
| 瓦埠湖 Wabuhu lake | 25 | 3 | 0.290±0.109 | 5 | 0.00072±0.00028 |
| 小计 Subtotal | 50 | 7 | 0.449±0.086 | 9 | 0.00071±0.00018 |
| 总体 All samples | 129 | 18 | 0.590±0.047 | 24 | 0.00088±0.00011 |
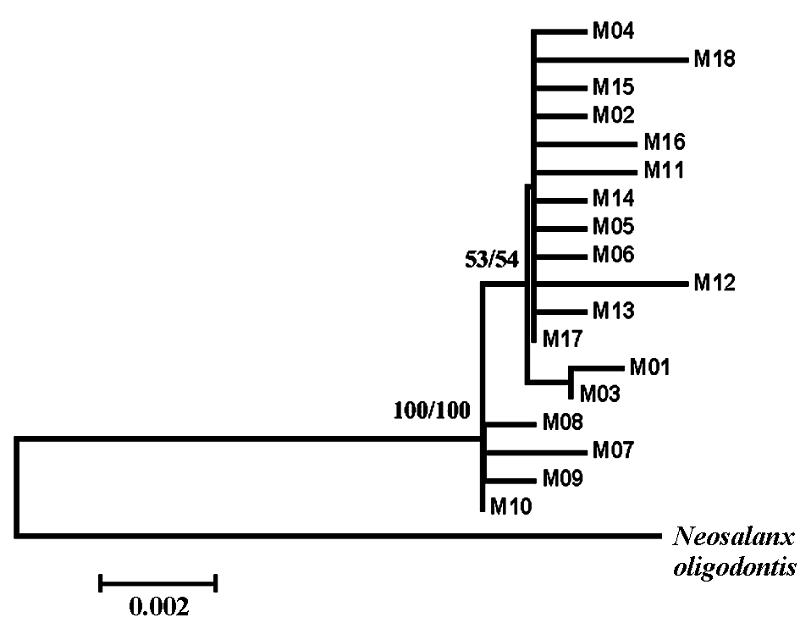
图1 邻接法和最大似然法构建的以寡齿新银鱼为外群的乔氏新银鱼18种单倍型系统进化树。枝的根部为bootstrap值。
Fig. 1 Neighbor-joining and Maximum-likelihood tree for all 18 haplotypes of Neosalanx jordani and for one outgroup taxa, N. oligodontis. Values indicate bootstrap support for each node.
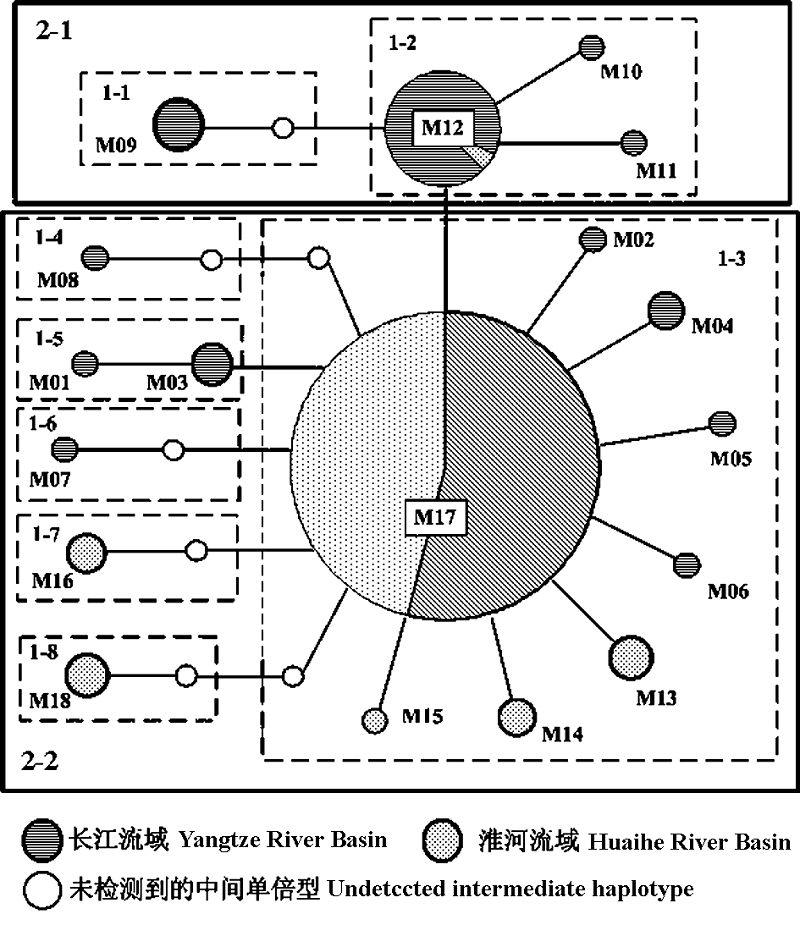
图2 使用TCS软件以最大简约法构建的最小跨度网络。图中每一个条线代表一个突变连接, 节点处的数字代表不同的单倍型, 节点大小对应单倍型出现频率。图中单倍型编号同表1, 白点代表未能在本研究中检测到的中间单倍型。围绕单倍型的方框确定了嵌套枝分析中的一步枝及两步枝。
Fig. 2 Minimum spanning network based on statistical parsimony using TCS software. Nodes indicate the haplotypes number and are proportional to the haplotype frequency. White nodes indicate undetected intermediate haplotype. Boxes indicate one-step to two-step nesting levels for the nested clade analysis.
| 变异来源 Source of variation | 自由度 d.f. | 平方和 Sum of squares | 变异组分 Variance components | 变异百分比 % variation | Ф-统计量 Ф-statistics |
|---|---|---|---|---|---|
| 流域间 Among basins | 1 | 4.491 | -0.0292 | -4.40 | ФCT =- 0.04396 |
| 流域内 Within basins | 3 | 18.961 | 0.2269 | 34.17 | ФSC= 0.3273** |
| 种群内 Within populations | 124 | 57.817 | 0.4663 | 70.22 | ФST = 0.2978** |
| 总和 Total | 128 | 81.270 | 0.6640 |
表3 乔氏新银鱼线粒体细胞色素b基于Ф-统计量的AMOVA分析结果
Table 3 Results of AMOVA forNeosalanx jordani mtDNA Cyt b estimation using Ф-statistics
| 变异来源 Source of variation | 自由度 d.f. | 平方和 Sum of squares | 变异组分 Variance components | 变异百分比 % variation | Ф-统计量 Ф-statistics |
|---|---|---|---|---|---|
| 流域间 Among basins | 1 | 4.491 | -0.0292 | -4.40 | ФCT =- 0.04396 |
| 流域内 Within basins | 3 | 18.961 | 0.2269 | 34.17 | ФSC= 0.3273** |
| 种群内 Within populations | 124 | 57.817 | 0.4663 | 70.22 | ФST = 0.2978** |
| 总和 Total | 128 | 81.270 | 0.6640 |
| 嵌套枝 Nested clade | P 值 Pvalue | 内部枝 Interior clades | 枝距离 Clade distance (DC) | 嵌套枝距离 Nested clade distance (DN) | 推断 Inference |
|---|---|---|---|---|---|
| Clade 1-3 | 0.000 | clade M02 (Tip) | 181.97 | 195.76 | 1-2-11RE-12-13-14 LDC/PF |
| clade M04 (Tip) | 0.00S | 249.12L | |||
| clade M05 (Tip) | 0.00 | 0.00 | |||
| clade M06 (Tip) | 0.00S | 178.79S | |||
| clade M13 (Tip) | 0.00S | 261.54L | |||
| clade M14 (Tip) | 89.68 S | 236.22 L | |||
| clade M15 (Tip) | 17.47S | 130.44S | |||
| clade M17 (Interior) | 19.87S | 194.75 | |||
| I-T | -3.78 | 0.00 | |||
| Clade 2-1 | 0.000 | clade 1-1 (Tip) | 247.12L | 281.70L | 1-2-11RE-12CRE |
| clade 1-2 (Interior) | 54.98S | 106.53S | |||
| I-T | -192.14S | -175.16S | |||
| Clade 2-2 | 0.000 | clade 1-3 (Interior) | 129.83S | 187.67 | 1-2-11RE-12CRE |
| clade 1-4 (Tip) | 44.50S | 176.87 | |||
| clade 1-5 (Tip) | 0.00S | 312.79L | |||
| clade 1-6 (Tip) | 0.00S | 196.43 | |||
| clade 1-7 (Tip) | 41.62S | 173.41 | |||
| clade 1-8 (Tip) | 79.46 | 173.26 | |||
| I-T | 98.96 | 0.96 | |||
| Total cladogram | 0.000 | clade 2-1 (Tip) | 239.66 | 227.23 | 1-2-11RE-12 CRE |
| clade 2-2 (Interior) | 0.00S | 136.82S | |||
| I-T | -241.43L | -1,100.14L |
表4 乔氏新银鱼线粒体细胞色素b单倍型嵌套枝分析结果
Table 4 Results of the nested clade analysis of Neosalanx jordani mtDNACyt b haplotypes
| 嵌套枝 Nested clade | P 值 Pvalue | 内部枝 Interior clades | 枝距离 Clade distance (DC) | 嵌套枝距离 Nested clade distance (DN) | 推断 Inference |
|---|---|---|---|---|---|
| Clade 1-3 | 0.000 | clade M02 (Tip) | 181.97 | 195.76 | 1-2-11RE-12-13-14 LDC/PF |
| clade M04 (Tip) | 0.00S | 249.12L | |||
| clade M05 (Tip) | 0.00 | 0.00 | |||
| clade M06 (Tip) | 0.00S | 178.79S | |||
| clade M13 (Tip) | 0.00S | 261.54L | |||
| clade M14 (Tip) | 89.68 S | 236.22 L | |||
| clade M15 (Tip) | 17.47S | 130.44S | |||
| clade M17 (Interior) | 19.87S | 194.75 | |||
| I-T | -3.78 | 0.00 | |||
| Clade 2-1 | 0.000 | clade 1-1 (Tip) | 247.12L | 281.70L | 1-2-11RE-12CRE |
| clade 1-2 (Interior) | 54.98S | 106.53S | |||
| I-T | -192.14S | -175.16S | |||
| Clade 2-2 | 0.000 | clade 1-3 (Interior) | 129.83S | 187.67 | 1-2-11RE-12CRE |
| clade 1-4 (Tip) | 44.50S | 176.87 | |||
| clade 1-5 (Tip) | 0.00S | 312.79L | |||
| clade 1-6 (Tip) | 0.00S | 196.43 | |||
| clade 1-7 (Tip) | 41.62S | 173.41 | |||
| clade 1-8 (Tip) | 79.46 | 173.26 | |||
| I-T | 98.96 | 0.96 | |||
| Total cladogram | 0.000 | clade 2-1 (Tip) | 239.66 | 227.23 | 1-2-11RE-12 CRE |
| clade 2-2 (Interior) | 0.00S | 136.82S | |||
| I-T | -241.43L | -1,100.14L |
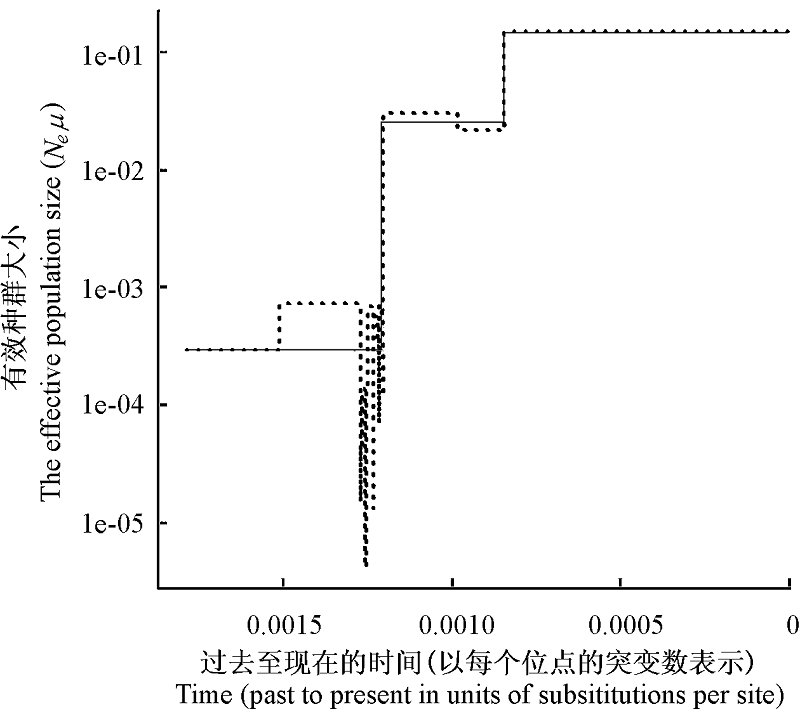
图3 有效种群大小随溯祖时间变化的Skyline图。虚线为Classic Skyline, 细实线为Generalized Skyline。
Fig. 3 Skyline plots of changes in effective population size on time. Classic Skyline was shown as dot line, and Generalized Skyline as slender line.
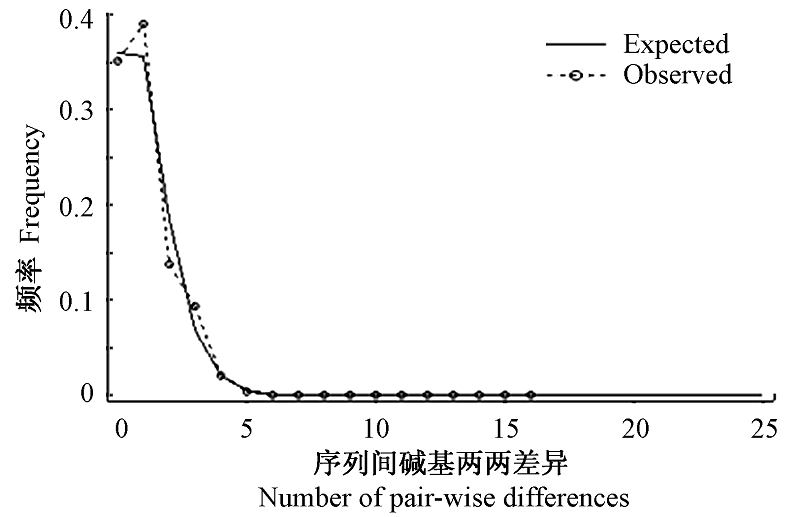
图4 歧点分布分析结果。虚线代表序列之间碱基两两差异的分布, 实线表示在扩张假设理论下歧点分布的期望曲线。
Fig. 4 Observed and expected mismatch distributions showing the frequencies of pair-wise differences. The observed pair-wise difference (dashed line), and the expected mismatch distributions under the sudden expansion model (solid line) ofCyt b.
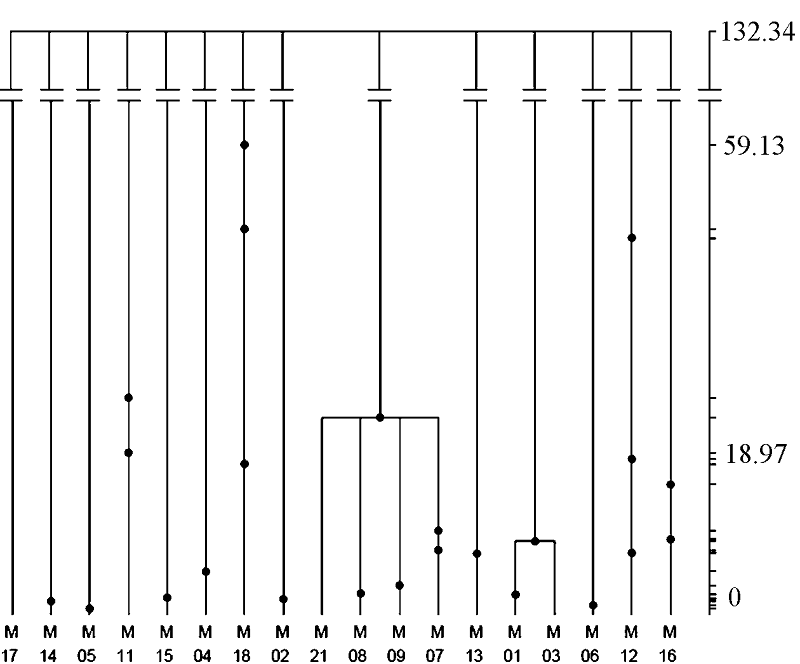
图5 乔氏新银鱼线粒体细胞色素b基因树。该基因树通过1,000,000 次溯祖模拟构建。右侧坐标为距现在的溯祖时间(单位为千年), 基因树上的点表示突变发生的历史时间。
Fig. 5 Gene tree of the Neosalanx jordani mitochondrialCyt bgene. The tree is based on 1,000,000 coalescent simulations. The right coordinate is scaled by absolute coalescent time (kyr), and the dots in the tree shows the ancestral distribution of mutations in the population history.
| [1] | Akaike H (1974) A new look at the statistical model identification. IEEE Transactions on Automatic Control, AC-19, 716-723. |
| [2] |
Avise JC (1989) A role for molecular genetics in the recognition and conservation of endangered species. Trends in Ecology and Evolution, 4, 279-281.
DOI URL PMID |
| [3] | Avise JC (2000) Phylogeography: the History and Formation of Species. Harvard University Press, Cambridge. |
| [4] |
Bahlo M, Griffiths RC (2000) Inference from gene trees in a subdivided population. Theoretical Population Biology, 57, 79-95.
URL PMID |
| [5] | Beheregaray LB, Ciofi C, Geist D, Gibbs JP, Caccone A, Powell JR (2003) Genes record a prehistoric volcanoe eruption in the Galapagos. Science, 302, 75. |
| [6] |
Buhay JE, Crandall KA (2005) Subterranean phylogeography of freshwater crayfishes shows extensive gene flow and surprisingly large population sizes. Molecular Ecology, 14, 4259-4273.
URL PMID |
| [7] |
Cantatore P, Roberti M, Pesole G, Ludovico A, Milella F, Gadaleta MN, Saccone C (1994) Evolutionary analysis of cytochrome b sequences in some Perciformes: evidence for a slower rate of evolution than in mammals. Journal of Molecular Evolution, 39, 589-597.
URL PMID |
| [8] | Cheng QT (成庆泰), Zheng BS (郑葆珊) (1987) Systematic Synopsis of Chinese Fishes (中国鱼类系统检索). Science Press, Beijing. (in Chinese) |
| [9] |
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Molecular Ecology, 9, 1657-1659.
URL PMID |
| [10] | Cook RM, Sinclair A, Stefansson G (1997) Potential collapse of North Sea cod stocks. Nature, 385, 521-522. |
| [11] |
Cortey M, Pla C, García-Marín JL (2004) Historical biogeography of Mediterranean trout. Molecular Phylogenetics and Evolution, 33, 831-844.
DOI URL PMID |
| [12] |
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics, 134, 959-969.
URL PMID |
| [13] | Currens KP, Schreck CB, Li WH (1990) Allozyme and morphological divergence of rainbow trout ( Oncorhynchus mykiss) above and below waterfalls in the Deschutes River, Oregon. Copeia, 1990, 730-746. |
| [14] | Emerson BC, Paradis E, Thebaud C (2001) Revealing the demographic histories of species using DNA sequences. Trends in Ecology and Evolution, 16, 707-716. |
| [15] |
Excoffier L, Laval LG, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47-50.
URL PMID |
| [16] | Fang P (1934a) Study on the fishes referring to Salangidae of China. Sinensia, 4, 231-268. |
| [17] | Fang P (1934b) Supplementary notes on the fishes referring to Salangidae of China. Sinensia, 5, 505-511. |
| [18] | Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conservation Biology, 10, 1500-1508. |
| [19] |
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915-925.
URL PMID |
| [20] | Grant WAS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89, 415-426. |
| [21] |
Harpending H (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology, 66, 591-600.
URL PMID |
| [22] |
Hashiguchi Y, Kado T, Kimura S, Tachida H (2006) Comparative phylogeography of two bitterlings, Tanakia lanceolata and T. limbata (Teleostei, Cyprinidae), in Kyushu and adjacent districts of western Japan, based on mitochondrial DNA analysis. Zoological Science, 23, 309-322.
URL PMID |
| [23] | Hynes R, Ferguson A, McCann M (1996) Variation in mitochondrial DNA and post-glacial colonisation of north-west Europe by brown trout ( Salmo trutta L.). Journal of Fish Biology, 48, 54-67. |
| [24] | Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. Journal of Computational and Graphical Statistics, 5, 299-314. |
| [25] | Inoue JG, Miya M, Tsukamoto K, Nishida M (2000) Complete mitochondrial DNA sequence of the Japanese sardine Sardinops melanostictus. Fisheries Science, 66, 924-932. |
| [26] |
Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, Beerli P, Su XZ (2003) Early origin and recent expansion of Plasmodium falciparum. Science, 300, 318-321.
URL PMID |
| [27] | Kamal MI, Richard AN, Godfrey MH (1996) Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity, 77, 282-291. |
| [28] |
Kuhner MK, Yamato J, Felsenstein J (1998) Maximum likelihood estimation of population growth rates based on the coalescent. Genetics, 149, 429-434.
URL PMID |
| [29] |
Lambeck K, Esat TM, Potter EK (2002) Links between climate and sea levels for the past three million years. Nature, 419, 199-206.
DOI URL PMID |
| [30] | Li N (李娜), Chen SB (陈少波), Xie QL (谢起浪), Lü JX (吕建新), Guan MX (管敏鑫) (2008) Polymorphisms of mitochondrial Cyt b gene and D-loop region in sweetfish ( Plecoglossus altivelis Temminck et Schlegel) from Zhejiang and Fujian provinces. Hereditas (Beijing) (遗传), 30, 919-925. (in Chinese with English abstract) |
| [31] |
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Research, 27, 209-220.
URL PMID |
| [32] | McCusker MR, Parkinson E, Taylor EB (2000) Mitochondrial DNA variation in rainbow trout ( Oncorhynchus mykiss) across its native range: testing biogeographical hypotheses and their relevance to conservation. Molecular Ecology, 9, 2089-2108. |
| [33] |
McGlashan DJ, Hughes JM (2000) Reconciling patterns of genetic variation with stream structure, earth history and biology in the Australian freshwater fish Craterocephalus stercusmuscarum (Atherinidae). Molecular Ecology, 9, 1737-1751.
DOI URL PMID |
| [34] | Moritz C (1994) Applications of mitochondrial DNA analysis in conservation: a critical review. Molecular Ecology, 3, 401-411. |
| [35] | Nelson J (1994) Fishes of the World, 3rd edn. John Wiley and Sons, Inc., New York. |
| [36] | Perdices A, Sayanda D, Coelho MM (2005) Mitochondrial diversity of Opsariichthys bidens (Teleostei, Cyprinidae) in three Chinese drainages. Molecular Phylogenetics and Evolution, 37, 920-927. |
| [37] |
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics, 14, 817-818.
DOI URL PMID |
| [38] |
Posada D, Crandall KA (2001) Intraspecific gene genealogies: trees grafting into networks. Trends in Ecology and Evolution, 16, 37-45.
DOI URL PMID |
| [39] |
Posada D, Crandall KA, Templeton AR (2000) GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Molecular Ecology, 9, 487-488.
URL PMID |
| [40] |
Pybus OG, Rambaut A, Harvey PH (2000) An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics, 155, 1429-1437.
URL PMID |
| [41] |
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution, 19, 2092-2100.
DOI URL PMID |
| [42] |
Riley SPD, Pollinger JP, Sauvajot RM, York EC, Bromley C, Fuller TK, Wayne RK (2006) A southern California freeway is a physical and social barrier to gene flow in carnivores. Molecular Ecology, 15, 1733-1741.
DOI URL PMID |
| [43] |
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9, 552-569.
DOI URL PMID |
| [44] |
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19, 2496-2497.
DOI URL PMID |
| [45] | Sheldon JM, Robert HD, Michael JS (1996) Phylogeny of Pacific salmon and trout based on growth hormone type-2 and mitochondrial NADH dehydrogenase subunit 3 DNA sequences. Canadian Journal of Fisheries and Aquatic Sciences, 53, 1165-1176. |
| [46] |
Sigurgislason H, Arnason E (2003) Extent of mitochondrial DNA sequence variation in Atlantic cod from the Faroe Islands: a resolution of gene genealogy. Heredity, 91, 557-564.
DOI URL PMID |
| [47] |
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics, 129, 555-562.
URL PMID |
| [48] | Smith GR (1992) Introgression in fishes: significance for paleontology, cladistics, and evolutionary rates. Systematic Biology, 41, 41-57. |
| [49] |
Strimmer K, Pybus OG (2001) Exploring the demographic history of DNA sequences using the generalized skyline plot. Molecular Biology and Evolution, 18, 2298-2305.
DOI URL PMID |
| [50] | Swofford D (2002) Paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts. |
| [51] |
Takehana Y, Uchiyama S, Matsuda M, Jeon SR, Sakaizumi M (2004) Geographic variation and diversity of the cytochrome b gene in wild populations of medaka ( Oryzias latipes) from Korea and China. Zoological Science, 21, 483-491.
DOI URL PMID |
| [52] |
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software Version 4.0. Molecular Biology and Evolution, 24, 1596-1599.
URL PMID |
| [53] |
Templeton AR (2004) Statistical phylogeography: methods of evaluating and minimizing inference errors. Molecular Ecology, 13, 789-809.
URL PMID |
| [54] |
Templeton AR, Boerwinkle E, Sing CF (1987) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics, 117, 343-351.
URL PMID |
| [55] |
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics, 132, 619-633.
URL PMID |
| [56] |
Templeton AR, Sing CF (1993) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics, 134, 659-669.
URL PMID |
| [57] | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. |
| [58] | Waits L (1989) Mitochondrial DNA phylogeography of the North American brown bear and implications for conservation. Conservation Biology, 13, 408-417. |
| [59] | Wang SM (王苏民), Dou HS (窦鸿身) (1998) The Lakes of China (中国湖泊志). Science Press, Beijing. (in Chinese) |
| [60] | Wang ZS (王忠锁), Fu CZ (傅萃长), Lei GC (雷光春) (2002) Biodiversity of Chinese icefishes (Salangidae) and their conserving strategies. Biodiversity Science (生物多样性), 10, 416-424. (in Chinese with English abstract) |
| [61] | Ward R, Woodwark M, Skibinski O (1994) A comparison of genetic diversity levels in marine, freshwater and anadromous fishes. Journal of Fish Biology, 44, 213-232. |
| [62] |
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theoretical Population Biology, 7, 256-276.
DOI URL PMID |
| [63] |
Whiteley AR, Spruell P, Allendorf FW (2006) Can common species provide valuable information for conservation? Molecular Ecology, 15, 2767-2786.
DOI URL PMID |
| [64] | Xia DQ (夏德全), Cao Y (曹萤), Wu TT (吴婷婷), Yang H (杨弘) (2000) Study on lineages of Protosalanx chinensis, Neosalanx taihuensis and N. oligodontis in Taihu Lake with RAPD technique. Journal of Fishery Sciences of China (中国水产科学), 7, 12-15. (in Chinese with English abstract) |
| [65] | Xia YZ, Chen YY, Sheng Y (2006) Phylogeographic structure of lenok ( Brachymystax lenok Pallas) (Salmoninae, Salmonidae) populations in water systems of eastern China, inferred from mitochondrial DNA sequences. Zoological Studies, 45, 190-200. |
| [66] | Zhang Y (张颖), Dong S (董仕), Wang Q (王茜), Sun ZR (孙振荣) (2005) The isozyme genetic structures in large icefish ( Protosalanx hyalocranius) and Taihu Lake icefish (Neosalanx taihuensis). Journal of Dalian Fisheries University (大连水产学院学报), 20, 111-115. (in Chinese with English abstract) |
| [67] | Zhang YL, Qiao XG (1994) Study on phylogeny and zoogeography of fishes of the family Salangidae. Acta Zoologica Taiwanica, 5, 95-115. |
| [68] | Zhao L, Zhang J, Liu Z, Funk S, Wei F, Xu M, Li M (2008) Complex population genetic and demographic history of the Salangid, Neosalanx taihuensis, based on cytochrome b sequences. BMC Evolutionary Biology, 8, 201. |
| [1] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [2] | 罗晓, 李峰, 陈静, 蒋志刚. 青海湖地区狗獾分类地位和狗獾属进化历史探讨[J]. 生物多样性, 2016, 24(6): 694-700. |
| [3] | 杜建国, WilliamW.L. Cheung, 陈彬, 周秋麟, 杨圣云, GuanqiongYe. 气候变化与海洋生物多样性关系研究进展[J]. 生物多样性, 2012, 20(6): 745-754. |
| [4] | 田瑜, 邬建国, 寇晓军, 李钟汶, 王天明, 牟溥, 葛剑平. 东北虎种群的时空动态及其原因分析[J]. 生物多样性, 2009, 17(3): 211-225. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn