



生物多样性 ›› 2022, Vol. 30 ›› Issue (6): 21545. DOI: 10.17520/biods.2021545 cstr: 32101.14.biods.2021545
李永光1,2, 任辉1, 张英杰1, 李瑞宁2, 艾昊2, 黄先忠2,3,*( )
)
收稿日期:2021-12-30
接受日期:2022-03-10
出版日期:2022-06-20
发布日期:2022-03-11
通讯作者:
黄先忠
作者简介:* E-mail: huangxz@ahstu.edu.cn基金资助:
Yongguang Li1,2, Hui Ren1, Yingjie Zhang1, Ruining Li2, Hao Ai2, Xianzhong Huang2,3,*( )
)
Received:2021-12-30
Accepted:2022-03-10
Online:2022-06-20
Published:2022-03-11
Contact:
Xianzhong Huang
摘要:
近年来植物基因组测序物种数量的指数增长, 为我们对植物环境适应性状的遗传和变异的全面理解提供了保障。磷脂酰乙醇胺结合蛋白(phosphatidylethanolamine-binding protein, PEBP)在植物的开花转变和株型建立中起着重要作用, 一直是植物生物学研究关注的热点领域之一。然而对该家族并没有利用新近测序的基因组数据进行比较基因组分析, 制约了对其在分子水平上的进化研究。为了确定PEBP基因家族的分子进化机制, 本研究利用生物信息学方法开展了7种十字花科植物拟南芥(Arabidopsis thaliana)、琴叶拟南芥(A. lyrata)、小鼠耳芥(A. pumila)、亚麻荠(Camelina sativa)、甘蓝(Brassica oleracea)、白菜(B. rapa)和油菜(B. napus)的PEBP基因家族成员的全基因组鉴定、结构特征和比较进化分析。从7个物种中共鉴定出91个PEBP基因, 系统进化分析表明它们分属5个亚家族: MFT、FT/TSF、TFL1、CEN和BFT。基因结构分析发现甘蓝、白菜和油菜的CEN基因内含子明显比其余4个物种的内含子长。蛋白结构域分析表明MFT比其他4个亚家族成员少了一个motif 2, TFL1比其他亚家族多了motif 8。选择压力分析发现7个物种PEBP同源基因均受到较强的纯化选择, 其中TFL1亚家族受到的纯化选择最弱。共线性分析表明十字花科植物PEBP基因家族随古代多倍体事件发生不同程度的扩张, TSF在甘蓝、白菜和油菜中丢失。非生物胁迫下, 在拟南芥中过量表达小鼠耳芥的一个MFT基因, 转基因拟南芥种子的萌发率明显低于野生型, 暗示MFT基因在调控种子萌发上的功能保守。本研究为深入研究十字花科植物PEBP基因的进化特征和生物学功能奠定了基础。
李永光, 任辉, 张英杰, 李瑞宁, 艾昊, 黄先忠 (2022) 十字花科植物PEBP基因家族的分子进化. 生物多样性, 30, 21545. DOI: 10.17520/biods.2021545.
Yongguang Li, Hui Ren, Yingjie Zhang, Ruining Li, Hao Ai, Xianzhong Huang (2022) Analysis of the molecular evolution of the PEBP gene family in cruciferous plants. Biodiversity Science, 30, 21545. DOI: 10.17520/biods.2021545.
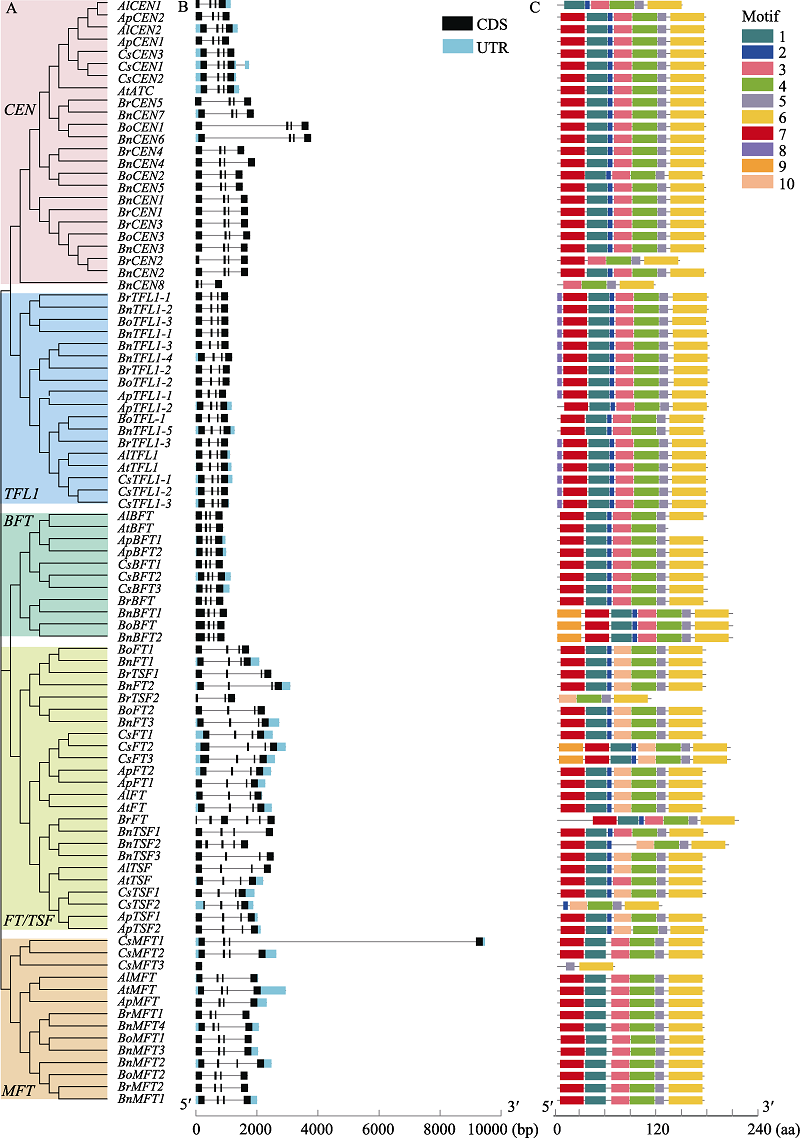
图1 拟南芥、琴叶拟南芥、小鼠耳芥、亚麻荠、甘蓝、白菜和油菜PEBP基因家族的基因结构及motif分布。A: 7个物种PEBP系统进化树; B: PEBP基因的外显子-内含子分布; C: PEBP蛋白保守基序分布特征。CDS: 蛋白质编码序列区域; UTR: 非编译区域; aa: 氨基酸。
Fig. 1 Motif distributions and exon-intron structures of the PEBP family members in Arabidopsis thaliana, A. lyrata, A. pumila, Camelina sativa, Brassica oleracea, B. rapa, and B. napus. A, PEBP phylogenetic tree of seven species; B, Exon-intron distribution of PEBP genes; C, Distribution characteristics of the conserved motifs of PEBP proteins; CDS, Coding sequence; UTR, Untranslated region; aa, Amino acid.
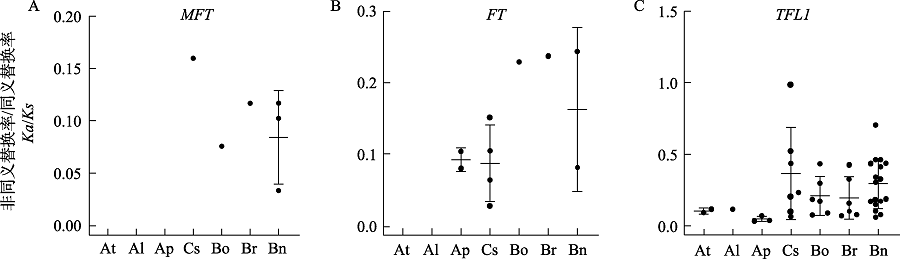
图2 物种内PEBP共线性基因对的选择压力统计。At: 拟南芥; Al: 琴叶拟南芥; Ap: 小鼠耳芥; Cs: 亚麻荠; Bo: 甘蓝; Br: 白菜; Bn: 油菜。
Fig. 2 Statistics of selection pressures on the PEBP collinearity genes within species. At, Arabidopsis thaliana; Al, A. lyrata; Ap, A. pumila; Cs, Camelina sativa; Bo, Brassica oleracea; Br, B. rapa; Bn, B. napus.
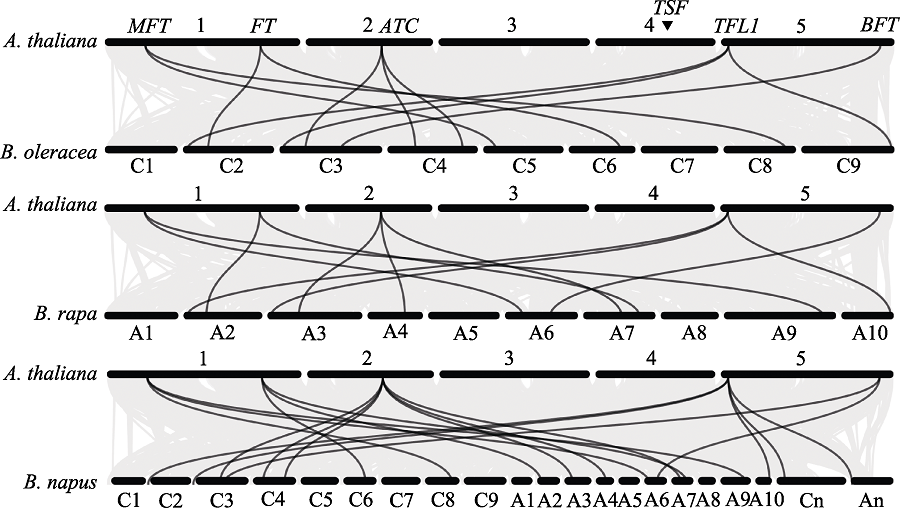
图3 拟南芥与甘蓝、白菜和油菜PEBP基因的共线性关系。圆柱分别代表拟南芥、甘蓝、白菜和油菜的5、9、10和21条染色体, 黑线表示物种间PEBP共线性关系。灰线表示基因组共线性关系。三角形代表TSF基因的位置。
Fig. 3 Syntenic relationships of PEBP genes in Arabidopsis thaliana, Brassica oleracea, B. rapa, and B. napus. The cylinder represents 5, 9, 10 and 21 chromosomes of A. thaliana, B. oleracea, B. rapa, and B. napus, respectively. The black line represents a common linear relationship of the PEBP gene, and the gray line represents a genomic linear relationship. The triangle represents the location of the TSF gene.
| 物种 Species | PEBP基因数量 No. of PEBP gene | 不同复制类型PEBP基因数量(百分比) No. of PEBP genes from different origins (percentage) | ||||
|---|---|---|---|---|---|---|
| 单例型 Singleton | 分散型 Dispersed | 近端型 Proximal | 串联重复 Tandem | 全基因组复制或片段复制 WGD/S | ||
| 拟南芥 Arabidopsis thaliana | 6 | 0 | 1 (16.6) | 0 | 0 | 5 (83.3) |
| 琴叶拟南芥 Arabidopsis lyrata | 7 | 0 | 7 (100) | 0 | 0 | 0 |
| 小鼠耳芥 Arabidopsis pumila | 11 | 0 | 1 (9.1) | 0 | 0 | 10 (90.9) |
| 亚麻芥 Camelina sativa | 17 | 0 | 1 (5.9) | 0 | 0 | 16 (94.1) |
| 甘蓝 Brassica oleracea | 11 | 0 | 1 (9.1) | 0 | 0 | 10 (90.9) |
| 白菜 Brassica rapa | 14 | 0 | 3 (21.4) | 1 (7.1) | 0 | 10 (71.4) |
| 油菜 Brassica napus | 25 | 0 | 6 (24) | 0 | 0 | 19 (76) |
表1 十字花科植物PEBP家族基因的复制类型
Table 1 Types of replication of PEBP gene family in Cruciferae plants
| 物种 Species | PEBP基因数量 No. of PEBP gene | 不同复制类型PEBP基因数量(百分比) No. of PEBP genes from different origins (percentage) | ||||
|---|---|---|---|---|---|---|
| 单例型 Singleton | 分散型 Dispersed | 近端型 Proximal | 串联重复 Tandem | 全基因组复制或片段复制 WGD/S | ||
| 拟南芥 Arabidopsis thaliana | 6 | 0 | 1 (16.6) | 0 | 0 | 5 (83.3) |
| 琴叶拟南芥 Arabidopsis lyrata | 7 | 0 | 7 (100) | 0 | 0 | 0 |
| 小鼠耳芥 Arabidopsis pumila | 11 | 0 | 1 (9.1) | 0 | 0 | 10 (90.9) |
| 亚麻芥 Camelina sativa | 17 | 0 | 1 (5.9) | 0 | 0 | 16 (94.1) |
| 甘蓝 Brassica oleracea | 11 | 0 | 1 (9.1) | 0 | 0 | 10 (90.9) |
| 白菜 Brassica rapa | 14 | 0 | 3 (21.4) | 1 (7.1) | 0 | 10 (71.4) |
| 油菜 Brassica napus | 25 | 0 | 6 (24) | 0 | 0 | 19 (76) |
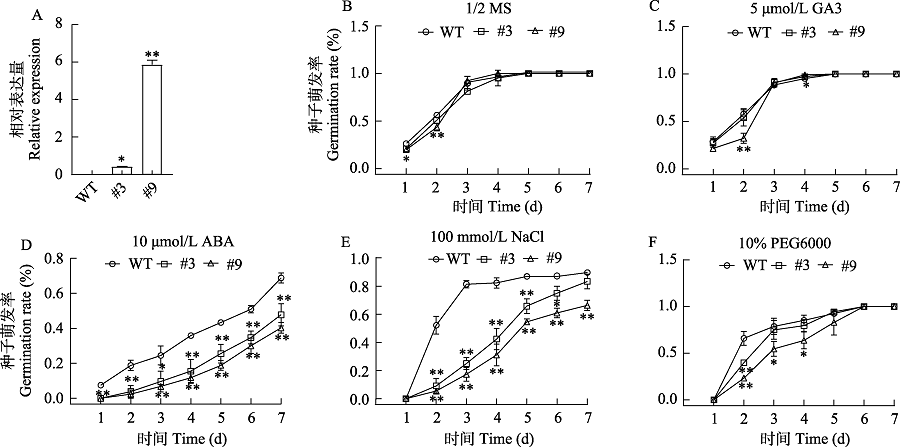
图4 35S:ApMFT转基因拟南芥在1/2 MS、ABA、GA3、NaCl、PEG6000处理下种子萌发率。WT: 野生型; #3和#9: 35S:ApMFT转基因株系; *差异显著(P < 0.05), **差异极显著(P < 0.01)。
Fig. 4 The germination rate of 35S:GhMFT1 transgenic Arabidopsis thaliana seeds treated with GA, ABA, NaCl and PEG6000. WT, Wild type; #3, #9: 35S:ApMFT transgenic lines, * Significant differences at P < 0.05; ** Significant differences at P < 0.01.
| [1] |
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science, 309, 1052-1056.
DOI URL |
| [2] |
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796-815.
DOI URL |
| [3] |
Banfield MJ, Barker JJ, Perry AC, Brady RL (1998) Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure, 6, 1245-1254.
PMID |
| [4] |
Bi ZH, Li X, Huang HS, Hua YW (2016) Identification, functional study, and promoter analysis of HbMFT1, a homolog of MFT from rubber tree (Hevea brasiliensis). International Journal of Molecular Sciences, 17, 247.
DOI URL |
| [5] |
Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Research, 13, 137-144.
DOI URL |
| [6] |
Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature, 379, 791-797.
DOI URL |
| [7] |
Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB, Wang XY, Chiquet J, Belcram H, Tong CB, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan GY, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan CC, Tollenaere R, Lu YH, Battail C, Shen JX, Sidebottom CHD, Wang XF, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu ZS, Sun FM, Lim YP, Lyons E, Town CD, Bancroft I, Wang XW, Meng JL, Ma JX, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou YM, Hua W, Sharpe AG, Paterson AH, Guan CY, Wincker P (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science, 345, 950-953.
DOI PMID |
| [8] |
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, Xia R (2020) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13, 1194-1202.
DOI URL |
| [9] |
Cheng SF, van den Bergh E, Zeng P, Zhong X, Xu JJ, Liu X, Hofberger J, de Bruijn S, Bhide AS, Kuelahoglu C, Bian C, Chen J, Fan GY, Kaufmann K, Hall JC, Becker A, Bräutigam A, Weber APM, Shi CC, Zheng ZJ, Li WJ, Lv MJ, Tao YM, Wang JY, Zou HF, Quan ZW, Hibberd JM, Zhang GY, Zhu XG, Xu X, Schranz ME (2013) The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell, 25, 2813-2830.
DOI URL |
| [10] |
Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal, 16, 735-743.
DOI URL |
| [11] |
Conti L, Bradley D (2007) TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell, 19, 767-778.
DOI URL |
| [12] |
Edger PP, Pires JC (2009) Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Research, 17, 699-717.
DOI URL |
| [13] |
Eshed Y, Lippman ZB (2019) Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science, 366, eaax0025.
DOI URL |
| [14] |
Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytologist, 183, 557-564.
DOI PMID |
| [15] |
Hedman H, Källman T, Lagercrantz U (2009) Early evolution of the MFT-like gene family in plants. Plant Molecular Biology, 70, 359-369.
DOI URL |
| [16] |
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G, Hollister JD, Ossowski S, Ottilar RP, Salamov AA, Schneeberger K, Spannagl M, Wang X, Yang L, Nasrallah ME, Bergelson J, Carrington JC, Gaut BS, Schmutz J, Mayer KFX, van de Peer Y, Grigoriev IV, Nordborg M, Weigel D, Guo YL (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics, 43, 476-481.
DOI URL |
| [17] |
Huang XZ, Yang LF, Jin YH, Lin J, Liu F (2017) Generation, annotation, and analysis of a large-scale expressed sequence tag library from Arabidopsis pumila to explore salt-responsive genes. Frontiers in Plant Science, 8, 955.
DOI URL |
| [18] | Jin S, Nasim Z, Susila H, Ahn JH (2021) Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Seminars in Cell & Developmental Biology, 109, 20-30. |
| [19] |
Jin YH, Liu F, Huang W, Sun Q, Huang XZ (2019) Identification of reliable reference genes for qRT-PCR in the ephemeral plant Arabidopsis pumila based on full-length transcriptome data. Scientific Reports, 9, 8408.
DOI URL |
| [20] |
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C, Higgins EE, Huebert T, Sharpe AG, Parkin IAP (2014) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nature Communications, 5, 3706.
DOI URL |
| [21] |
Karlgren A, Gyllenstrand N, Källman T, Sundström JF, Moore D, Lascoux M, Lagercrantz U (2011) Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution. Plant Physiology, 156, 1967-1977.
DOI PMID |
| [22] |
Li Q, Fan CM, Zhang XM, Wang X, Wu FQ, Hu RB, Fu YF (2014) Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS ONE, 9, e99642.
DOI URL |
| [23] | Li XC, Kang KC, Huang XZ, Fan YB, Song MM, Huang YJ, Ding JJ (2020) Genome-wide identification, phylogenetic analysis and expression profiling of the MKK gene family in Arabidopsis pumila. Hereditas (Beijing), 42, 403-421. (in Chinese with English abstract) |
| [李晓翠, 康凯程, 黄先忠, 范永斌, 宋苗苗, 黄韵杰, 丁佳佳 (2020) 小拟南芥MKK基因家族全基因组鉴定及进化和表达分析. 遗传, 42, 403-421.] | |
| [24] |
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IAP, Zhao M, Ma J, Yu J, Huang S, Wang X, Wang J, Lu K, Fang Z, Bancroft I, Yang TJ, Hu Q, Wang X, Yue Z, Li H, Yang L, Wu J, Zhou Q, Wang W, King GJ, Pires JC, Lu C, Wu Z, Sampath P, Wang Z, Guo H, Pan S, Yang L, Min J, Zhang D, Jin D, Li W, Belcram H, Tu J, Guan M, Qi C, Du D, Li J, Jiang L, Batley J, Sharpe AG, Park BS, Ruperao P, Cheng F, Waminal NE, Huang Y, Dong C, Wang L, Li J, Hu Z, Zhuang M, Huang Y, Huang J, Shi J, Mei D, Liu J, Lee TH, Wang J, Jin H, Li Z, Li X, Zhang J, Xiao L, Zhou Y, Liu Z, Liu X, Qin R, Tang X, Liu W, Wang Y, Zhang Y, Lee J, Kim HH, Denoeud F, Xu X, Liang X, Hua W, Wang X, Wang J, Chalhoub B, Paterson AH (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature Communications, 5, 3930.
DOI URL |
| [25] |
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25, 402-408.
DOI PMID |
| [26] |
Marks RA, Hotaling S, Frandsen PB, VanBuren R (2021) Representation and participation across 20 years of plant genome sequencing. Nature Plants, 7, 1571-1578.
DOI PMID |
| [27] |
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell, 23, 3215-3229.
DOI URL |
| [28] |
Qiao X, Li M, Li LT, Yin H, Wu JY, Zhang SL (2015) Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biology, 15, 12.
DOI PMID |
| [29] |
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution, 34, 3299-3302.
DOI URL |
| [30] |
Song XM, Ma X, Li CJ, Hu JJ, Yang QH, Wang T, Wang L, Wang JP, Guo D, Ge WN, Wang ZY, Li MM, Wang QM, Ren TZ, Feng SY, Wang LX, Zhang WM, Wang XY (2018) Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genomics, 19, 346.
DOI URL |
| [31] |
Sun YQ, Shang LG, Zhu QH, Fan LJ, Guo LB (2022) Twenty years of plant genome sequencing: Achievements and challenges. Trends in Plant Science, 27, 391-401.
DOI URL |
| [32] |
Susila H, Jurić S, Liu L, Gawarecka K, Chung KS, Jin S, Kim SJ, Nasim Z, Youn G, Suh MC, Yu H, Ahn JH (2021) Florigen sequestration in cellular membranes modulates temperature-responsive flowering. Science, 373, 1137-1142.
DOI URL |
| [33] |
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science, 316, 1033-1036.
DOI URL |
| [34] |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725-2729.
DOI URL |
| [35] |
Taoka KI, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature, 476, 332-335.
DOI URL |
| [36] |
Thomas BC, Pedersen B, Freeling M (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Research, 16, 934-946.
PMID |
| [37] | Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics, Chapter 2, Unit 2. 3. |
| [38] | Wang XY, Gowik U, Tang HB, Bowers JE, Westhoff P, Paterson AH (2009) Comparative genomic analysis of C 4 photosynthetic pathway evolution in grasses. Genome Biology, 10, R68. |
| [39] |
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IAP, Batley J, Kim JS, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z (2011) The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics, 43, 1035-1039.
DOI URL |
| [40] |
Wang YP, Tang HB, Debarry JD, Tan X, Li JP, Wang XY, Lee TH, Jin HZ, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research, 40, e49.
DOI URL |
| [41] |
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science, 309, 1056-1059.
DOI URL |
| [42] |
Xi WY, Liu C, Hou XL, Yu H (2010) MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell, 22, 1733-1748.
DOI URL |
| [43] | Yadav CB, Bonthala VS, Muthamilarasan M, Pandey G, Khan Y, Prasad M (2015) Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Research, 22, 79-90. |
| [44] |
Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant and Cell Physiology, 46, 1175-1189.
PMID |
| [45] |
Yu XL, Liu H, Sang N, Li YF, Zhang TT, Sun J, Huang XZ (2019) Identification of cotton MOTHER OF FT AND TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination. PLoS ONE, 14, e0215771.
DOI URL |
| [46] |
Zhang B, Li C, Li Y, Yu H (2020) Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nature Plants, 6, 1146-1157.
DOI PMID |
| [47] |
Zhang J, Peng HW, Xia FC, Wang W (2021) A comparison of seed plants’ polyploids between the Qinghai-Tibet Plateau alpine and the Pan-Arctic regions. Biodiversity Science, 29, 1470-1480. (in Chinese with English abstract)
DOI |
|
[张军, 彭焕文, 夏富才, 王伟 (2021) 青藏高原高山区和泛北极地区种子植物多倍体比较. 生物多样性, 29, 1470-1480.]
DOI |
|
| [48] |
Zhou HS, Qi KJ, Liu X, Yin H, Wang P, Chen JQ, Wu JY, Zhang SL (2016) Genome-wide identification and comparative analysis of the cation proton antiporters family in pear and four other Rosaceae species. Molecular Genetics and Genomics, 291, 1727-1742.
DOI URL |
| [1] | 胡德美, 姚仁秀, 陈燕, 游贤松, 王顺雨, 汤晓辛, 王晓月. 青篱柴通过促进亲和花粉生长而提高传粉精确性[J]. 生物多样性, 2021, 29(7): 887-896. |
| [2] | 王爱霞, 马婧婧, 龚会蝶, 范国安, 王茂, 赵红梅, 程军回. 北疆一年生早春短命植物物种丰富度分布格局及其影响因素[J]. 生物多样性, 2021, 29(6): 735-745. |
| [3] | 李晶, 周天阳, 鲁雪丽, 李新涛, 孙斌, 孟红杰. 珍稀植物连香树在其中国分布区北缘的种子性状及幼苗更新限制[J]. 生物多样性, 2020, 28(10): 1161-1173. |
| [4] | 王晓月,朱鑫鑫,杨娟,刘云静,汤晓辛. 梅花个体内花柱长度的变异及其对繁殖成功的影响[J]. 生物多样性, 2019, 27(2): 159-167. |
| [5] | 张洋, 谭敦炎. 地下结实植物白番红花的繁育系统与传粉生物学[J]. 生物多样性, 2009, 17(5): 468-475. |
| [6] | 唐海萍, 颜莉娟, 张新时. 新疆准噶尔盆地生物多样性保育与建立国家荒漠公园的构想[J]. 生物多样性, 2008, 16(6): 618-626. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn