



Biodiv Sci ›› 2010, Vol. 18 ›› Issue (2): 129-136. DOI: 10.3724/SP.J.1003.2010.134 cstr: 32101.14.SP.J.1003.2010.134
Previous Articles Next Articles
Yan Liu1,2, Daoyuan Zhang1,*( )
)
Received:2009-07-07
Accepted:2009-11-16
Online:2010-03-20
Published:2010-03-20
Contact:
Daoyuan Zhang
Yan Liu, Daoyuan Zhang. Spatial genetic structure in five natural populations of Eremosparton songoricum as revealed by ISSR analysis[J]. Biodiv Sci, 2010, 18(2): 129-136.
| 居群 Population | 经纬度 Location | 海拔(m) Altitude | 群丛类型 Associatioin type | 生境 Habitat |
|---|---|---|---|---|
| A | 44°38'N 88°16'E | 548 | 准噶尔无叶豆 + 蛇麻黄群丛 Eremosparton songoricum+ Ephedra distachyaassociation | 南北向沙垄下部风蚀流沙地(位于沙漠腹地) Wind erosion sand surface of the lower south-north dune (located in hinterland desert, small population) |
| B | 44°46'N 88°16'E | 605 | 准噶尔无叶豆 + 沙蒿 + 三芒草群丛 Eremosparton songoricum + Artemisia arenaria + Aristida pennataassociation | 垄间低地及沙丘中下部(位于沙漠腹地) Interdune and middle-lower part of the dune (located in hinterland desert) |
| D | 44°58'N 88°25'E | 647 | 准噶尔无叶豆群丛 Eremosparton songoricumassociation | 道路和引水工程等建设区周围或两侧的缓起伏沙垄(沙丘)上(位于沙漠腹地) On dunes beside the road or encircle the transferring water project (located in hinterland desert) |
| F | 44°14'N 90°04'E | 718 | 准噶尔无叶豆 + 三芒草 + 一年生长营养期植物群系丛 Eremosparton songoricum + Aristida pennata + annual plant association | 垄底部至缓起伏的垄顶(位于沙漠东缘) From the bottom to the top of dune (located in east of desert) |
| G | 46°21'N 88°38'E | 794 | 准噶尔无叶豆 + 沙蒿群丛 Eremosparton songoricum + Artemisia arenaria association | 平缓流沙地(位于沙漠北缘, 紧邻乌仑谷湖) Flat wind erosion sand dune (located in north of desert, beside Wulungu Lake) |
Table 1 Distribution, association type and habitats of five Eremosparton songoricum populations in Gurbantunggut Desert
| 居群 Population | 经纬度 Location | 海拔(m) Altitude | 群丛类型 Associatioin type | 生境 Habitat |
|---|---|---|---|---|
| A | 44°38'N 88°16'E | 548 | 准噶尔无叶豆 + 蛇麻黄群丛 Eremosparton songoricum+ Ephedra distachyaassociation | 南北向沙垄下部风蚀流沙地(位于沙漠腹地) Wind erosion sand surface of the lower south-north dune (located in hinterland desert, small population) |
| B | 44°46'N 88°16'E | 605 | 准噶尔无叶豆 + 沙蒿 + 三芒草群丛 Eremosparton songoricum + Artemisia arenaria + Aristida pennataassociation | 垄间低地及沙丘中下部(位于沙漠腹地) Interdune and middle-lower part of the dune (located in hinterland desert) |
| D | 44°58'N 88°25'E | 647 | 准噶尔无叶豆群丛 Eremosparton songoricumassociation | 道路和引水工程等建设区周围或两侧的缓起伏沙垄(沙丘)上(位于沙漠腹地) On dunes beside the road or encircle the transferring water project (located in hinterland desert) |
| F | 44°14'N 90°04'E | 718 | 准噶尔无叶豆 + 三芒草 + 一年生长营养期植物群系丛 Eremosparton songoricum + Aristida pennata + annual plant association | 垄底部至缓起伏的垄顶(位于沙漠东缘) From the bottom to the top of dune (located in east of desert) |
| G | 46°21'N 88°38'E | 794 | 准噶尔无叶豆 + 沙蒿群丛 Eremosparton songoricum + Artemisia arenaria association | 平缓流沙地(位于沙漠北缘, 紧邻乌仑谷湖) Flat wind erosion sand dune (located in north of desert, beside Wulungu Lake) |
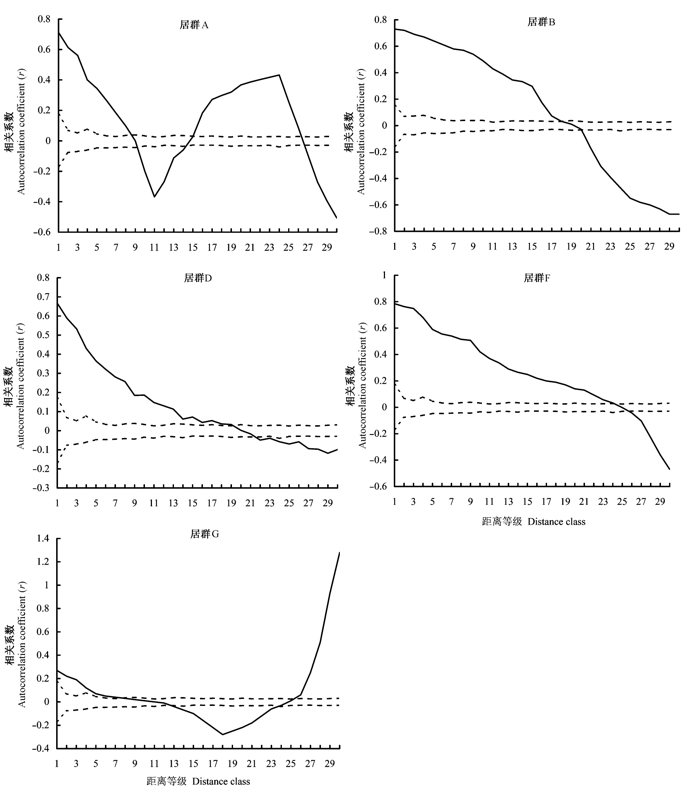
Fig. 1 Correlogram using mean value of Moran’sIof ISSR loci frequency variance for five Eremosparton songoricum populations. Solid lines represent the correlation coefficient (r) and dashed lines represent the 95% confidence intervals.
| 距离等级 Distance class | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1.0 | 2.0 | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 | 8.0 | 9.0 | 10.0 | |
| 居群A Population A | ||||||||||
| 正相关(%) | 31(93.9) | 19(57.6) | 4(12.1) | 6(18.2) | 11(33.3) | 19(57.6) | 19(57.6) | 24(72.7) | 8(24.2) | 2(6.1) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 13(39.4) | 17(51.5) | 18(54.5) | 16(48.5) | 11(33.3) | 8(24.2) | 7(21.2) | 21(63.6) | 26(78.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 93.9 | 97.0 | 63.6 | 72.7 | 81.8 | 90.9 | 81.8 | 93.9 | 87.9 | 84.8 |
| Significant correlation (%) | ||||||||||
| 居群B Population B | ||||||||||
| 正相关(%) | 25(100) | 19(76.0) | 17(68.0) | 15(60.0) | 16(64.0) | 12(48.0) | 8(32.0) | 3(12.0) | 2(8.0) | 1(4.0) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 1(4.0) | 4(16.0) | 7(28.0) | 8(32.0) | 10(40.0) | 17(68.0) | 20(80.0) | 23(92.0) | 21(84.0) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 80.0 | 84.0 | 88.0 | 96.0 | 88.0 | 100.0 | 92.0 | 100.0 | 88.0 |
| Significant correlation (%) | ||||||||||
| 居群D Population D | ||||||||||
| 正相关(%) | 27(100) | 23(85.2) | 16(59.3) | 14(51.9) | 14(51.9) | 13(48.1) | 10(37.0) | 8(29.6) | 5(18.5) | 6(22.2) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 3(11.1) | 9(33.3) | 13(48.1) | 12(44.4) | 12(44.4) | 15(55.6) | 18(66.7) | 19(70.4) | 20(74.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 96.3 | 92.6 | 100.0 | 96.3 | 92.6 | 92.6 | 96.3 | 88.9 | 96.3 |
| Significant correlation (%) | ||||||||||
| 居群F Population F | ||||||||||
| 正相关(%) | 28(100) | 19(67.9) | 20(71.4) | 15(53.6) | 17(60.7) | 11(39.3) | 15(53.6) | 14(50.0) | 10(35.7) | 4(14.3) |
| Positive correlation (%) | ||||||||||
| 负相关(比例) | 0(0) | 4(14.3) | 7(25.0) | 8(28.6) | 10(35.7) | 8(28.6) | 11(39.3) | 12(42.9) | 18(64.3) | 23(82.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 82.1 | 96.4 | 82.1 | 96.4 | 67.9 | 92.9 | 92.9 | 100.0 | 96.4 |
| Significant correlation (%) | ||||||||||
| 居群G Population G | ||||||||||
| 正相关(%) | 22(75.9) | 14(48.3) | 7(24.1) | 11(37.9) | 9(31.0) | 7(24.1) | 9(31.0) | 10(34.5) | 21(72.4) | 24(82.8) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 9(31.0) | 5(17.2) | 12(41.4) | 14(48.3) | 18(62.1) | 15(51.7) | 12(41.4) | 3(10.3) | 2(6.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 75.9 | 79.3 | 41.4 | 79.3 | 79.3 | 86.2 | 82.8 | 75.9 | 82.8 | 89.7 |
| Significant correlation (%) | ||||||||||
Table 2 Number of loci showing significant correlation in each distance class for Eremosparton songoricum in different populations (equal distance interval) and the percentage
| 距离等级 Distance class | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1.0 | 2.0 | 3.0 | 4.0 | 5.0 | 6.0 | 7.0 | 8.0 | 9.0 | 10.0 | |
| 居群A Population A | ||||||||||
| 正相关(%) | 31(93.9) | 19(57.6) | 4(12.1) | 6(18.2) | 11(33.3) | 19(57.6) | 19(57.6) | 24(72.7) | 8(24.2) | 2(6.1) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 13(39.4) | 17(51.5) | 18(54.5) | 16(48.5) | 11(33.3) | 8(24.2) | 7(21.2) | 21(63.6) | 26(78.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 93.9 | 97.0 | 63.6 | 72.7 | 81.8 | 90.9 | 81.8 | 93.9 | 87.9 | 84.8 |
| Significant correlation (%) | ||||||||||
| 居群B Population B | ||||||||||
| 正相关(%) | 25(100) | 19(76.0) | 17(68.0) | 15(60.0) | 16(64.0) | 12(48.0) | 8(32.0) | 3(12.0) | 2(8.0) | 1(4.0) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 1(4.0) | 4(16.0) | 7(28.0) | 8(32.0) | 10(40.0) | 17(68.0) | 20(80.0) | 23(92.0) | 21(84.0) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 80.0 | 84.0 | 88.0 | 96.0 | 88.0 | 100.0 | 92.0 | 100.0 | 88.0 |
| Significant correlation (%) | ||||||||||
| 居群D Population D | ||||||||||
| 正相关(%) | 27(100) | 23(85.2) | 16(59.3) | 14(51.9) | 14(51.9) | 13(48.1) | 10(37.0) | 8(29.6) | 5(18.5) | 6(22.2) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 3(11.1) | 9(33.3) | 13(48.1) | 12(44.4) | 12(44.4) | 15(55.6) | 18(66.7) | 19(70.4) | 20(74.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 96.3 | 92.6 | 100.0 | 96.3 | 92.6 | 92.6 | 96.3 | 88.9 | 96.3 |
| Significant correlation (%) | ||||||||||
| 居群F Population F | ||||||||||
| 正相关(%) | 28(100) | 19(67.9) | 20(71.4) | 15(53.6) | 17(60.7) | 11(39.3) | 15(53.6) | 14(50.0) | 10(35.7) | 4(14.3) |
| Positive correlation (%) | ||||||||||
| 负相关(比例) | 0(0) | 4(14.3) | 7(25.0) | 8(28.6) | 10(35.7) | 8(28.6) | 11(39.3) | 12(42.9) | 18(64.3) | 23(82.1) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 100.0 | 82.1 | 96.4 | 82.1 | 96.4 | 67.9 | 92.9 | 92.9 | 100.0 | 96.4 |
| Significant correlation (%) | ||||||||||
| 居群G Population G | ||||||||||
| 正相关(%) | 22(75.9) | 14(48.3) | 7(24.1) | 11(37.9) | 9(31.0) | 7(24.1) | 9(31.0) | 10(34.5) | 21(72.4) | 24(82.8) |
| Positive correlation (%) | ||||||||||
| 负相关(%) | 0(0) | 9(31.0) | 5(17.2) | 12(41.4) | 14(48.3) | 18(62.1) | 15(51.7) | 12(41.4) | 3(10.3) | 2(6.9) |
| Negative correlation (%) | ||||||||||
| 显著相关比例 | 75.9 | 79.3 | 41.4 | 79.3 | 79.3 | 86.2 | 82.8 | 75.9 | 82.8 | 89.7 |
| Significant correlation (%) | ||||||||||
| [1] | Васипьченко (1941) ФЛОРА СССР. Tomus Ⅺ (苏联植物志(第十一卷)), pp.310-311. (in Russian). |
| [2] |
Charlesworth B, Langley CH (1989) The population genetics of Drosophila transposable elements. Annual Review of Genetics, 23,251-287.
DOI URL PMID |
| [3] | Chen CD (陈昌笃), Zhang LY (张立运), Hu WK (胡文康) (1983) The basic characteristics of plant communities, flora and their distribution in the sandy district of Gurbantunggut. Acta Phytoecologica et Geobotanica Sinica (植物生态学与地植物学丛刊), 7,89-99. (in Chinese with English abstract) |
| [4] |
Chung MG, Chung JM, Chung MY, Epperson BK (2000) Spatial genetic distribution of allozyme polymorphisms following clonal and sexual reproduction in populations of Rhus javanica (Anacardiaceae) . Heredity, 84,178-185.
URL PMID |
| [5] |
Chung MY, Nason JD, Chung MG (2004) Spatial genetic structure in populations of the terrestrial orchid Cephalanthera longibracteata (Orchidaceae) . American Journal of Botany, 91,52-57.
DOI URL PMID |
| [6] |
Dong M (1995) Morphological responses to local light conditions of clonal herbs from contrasting habitats, and their modification due to physiological integration. Oecologia, 101,282-288.
DOI URL PMID |
| [7] | Eckert CG (2000) Contributions of autogamy and geitonogamy to self-fertilization in a mass-flowering, clonal plant. Ecology, 81,532-542. |
| [8] | Eckert CG, Barrett SCH (1993) Clonal reproduction and patterns of genotypic diversity in Decodon verticillatus (Lythraceae) . American Journal of Botany, 80,1175-1182. |
| [9] | Eckert CG (2002) The loss of sex in clonal plants. Evolutionary Ecology, 15,501-520. |
| [10] | Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics, 24,2l7-242. |
| [11] |
Epperson BK (1990) Spatial autocorrelation of genotypes under directional selection. Genetics, 124,757-771.
URL PMID |
| [12] |
Eriksson O (1993) Dynamics of genets in clonal plants. Trends in Ecology and Evolution, 8,313-316.
DOI URL PMID |
| [13] | Falk DA, Holsinger KE (1991) Genetics and Conservation of Rare Plants. Oxford University Press, Oxford, |
| [14] | Geburek T, Tripp-Knowles P (1994) Genetic architecture in buroak, Quercus macrocarpa (Fagaceae), inferred by means of spatial autocorrelation analysis, Plant Systematics and Evolution, 189,63-74. |
| [15] | Hamrick JL, Godt MJW, Murawski DA, Loveless MD (1991) Correlations between species traits and allozyme diversity:implications for conservation biology. In: In: Genetics and Conservation of Rare Plants (eds Falk DA, Holsinger KE)pp.75-86. Oxford University Press, New York. |
| [16] | Handel SN (1985) The intrusion of clonal growth patterns of plant breeding systems. The American Naturalist, 125,367-384. |
| [17] | Huenneke LF (1991) Ecological implications of genetic variation in plant populations. In: In: Genetics and Conservation of Rare Plants (eds Falk DA, Holsinger KE), pp.31-44. Oxford University Press, New York. |
| [18] |
Kelly BA, Hardy OJ, Bouvet JM (2004) Temporal and spatial genetic structure in Vitellaria paradoxa (shea tree) in an agroforestry system in southern Mali . Molecular Ecology, 13,1231-1240.
DOI URL PMID |
| [19] |
Lande R (1988) Genetics and demography in biological conservation. Science, 241,1455-1460.
DOI URL PMID |
| [20] | Leberg PL (1990) Genetic considerations in the design of introduction programs. Transactions of the North American Wildlife and Natural Resource Conference, 55,609-619. |
| [21] |
Levin DA, Kerster HW (1974) Gene flow in seed plants. Evolutionary Biology, 7,139-220.
DOI URL PMID |
| [22] | Li ZZ (李作洲), Lang P (郎萍), Huang HW (黄宏文) (2002) Spatial structure of allozyme frequencies in Castanea mollissima BI. Journal of Wuhan Botanical Research (武汉植物学研究), 20,165-170. (in Chinese with English abstract) |
| [23] | Li ZZ (李作洲), Gong JJ (龚俊杰), Wang Y (王瑛), Huang HW (黄宏文) (2003) Spatial structure of AFLP genetic diversity of remnant populations of Metasequoia glyptostroboides (Taxodiaceae) . Biodiversity Science (生物多样性), 11,265-275. (in Chinese with English abstract) |
| [24] | Liu YL (刘亚令), Li ZZ (李作洲), Zhang PF (张鹏飞), Jiang ZW (姜正旺), Huang HW (黄宏文) (2006) Spatial genetic structure in natural populations of two closely related Actinidia species (Actinidiaceae) as revealed by SSR analysis . Biodiversity Science (生物多样性), 14,421-434. (in Chinese with English abstract) |
| [25] | Loiselle BA, Sork VL, Nason J, Grnham C (1995) Spatial genetic structure of a tropical understory shrub. Psychotria offinalis (Rubiaceae). American Journal of Botany, 82,1420-1425. |
| [26] | Lu XY (陆雪莹), Zhang DY (张道远), Ma WB (马文宝) (2007) Genetic variation and clonal diversity in fragmented populations of the desert plant Eremosparton songoricum based on ISSR markers . Biodiversity Science (生物多样性), 15,282-291. (in Chinese with English abstract) |
| [27] | Ma WB (马文宝), Shi X (施翔), Zhang DY (张道远), Yin LK (尹林克) (2008) Flowering phenology and reproductive features of the rare plant Eremosparton songoricum in desert zone, Xinjiang, China . Journal of Plant Ecology (植物生态学报), 32,760-767. (in Chinese with English abstract) |
| [28] | Ma WB (马文宝), Zhang DY (张道远), Yin LK (尹林克) (2007) Variation of seed sizes of Eremosparton songoricum geographic populations and germination characteristics . Arid Land Geography (干旱区地理), 30,674-679. (in Chinese with English abstract) |
| [29] |
Marquardt PE, Epperson BK (2004) Spatial and population genetic structure of microsatellites in white pine. Molecular Ecology, 13,3305-33l5.
DOI URL PMID |
| [30] | Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6,288-295. |
| [31] | Shi X, Wang JC, Zhang DY, Gaskin J, Pan BR (2010a) Pollination ecology of the rare desert species Eremosparton songoricum (Fabaceae) . Australian Journal of Botany, 58,35-41. |
| [32] | Shi X, Wang JC, Zhang DY, Gaskin J, Pan BR (2010b) Pollen source and resource limitation to fruit production in the rare species Eremosparton songoricum (Fabaceae) . Nordic Journal of Botany, 28,1-7. |
| [33] |
Sokal RR, Jacquez GM, Wooten M (1989) Spatial autocorrelation analysis of migration and selection. Genetics, 121,845-855.
URL PMID |
| [34] | Sokal RR, Oden NL (1978) Spatial autocorrelation in biology. 1. Methodology. Biological Journal of the Linnean Society, 10,199-228. |
| [35] | Sutherland S, Delph LF (1984) On the importance of male fitness in plants: patterns of fruit-set. Ecology, 65,1093-1104. |
| [36] | Wang JC (王建成), Shi X (施翔), Zhang DY (张道远), Yin LK (尹林克) (2009) The morphological plasticity of Eremosparton songoricum along heterogeneous micro-habitats of continuous moisture gradient changes in sand duns . Acta Ecologica Sinica (生态学报), 29,3641-3648. (in Chinese with English abstract) |
| [37] | Warternberg D (1989) SAAP 4.3: A Spatial Autocorrelation Analysis Program.. Exeter Software, Setauket, New York. |
| [38] | Yin LK (尹林克), Tan LX (谭丽霞), Wang B (王兵) (2006) Rare and Endangered Higher Plants Endemic to Xinjiang (新疆珍稀濒危特有高等植物), pp.74-75. Xinjiang Scientific and Technical Publishing House, Urumq. (in Chinese) |
| [39] |
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution, 11,413-418.
DOI URL PMID |
| [40] | Zhang DY (张道远), Ma WB (马文宝), Shi X (施翔), Wang JC (王建成), Wang XY (王习勇) (2008) Distribution and bio-ecological characteristics of Eremosparton songoricum, a rare plant in Gurbantunggut desert . Journal of Desert Research (中国沙漠), 28,430-436. (in Chinese with English abstract) |
| [41] | Zhang DY (张道远), Wang JC (王建成), Shi X (施翔) (2009) Population quantitative dynamics of the rhizomatous woody clonal plant Eremosparton songoricum in China’s Gurbantunggut Desert . Chinese Journal of Plant Ecology (植物生态学报), 33,893-900. (in Chinese with English abstract) |
| [42] | Zhang LY (张立运), Hai Y (海鹰) (2002) Plant communities excluded in the book of “ The Vegetation and Its Utilization in Xinjiang”.Ⅰ. The desert plant communities . Arid Land Geography (干旱区地理), 25,84-89. (in Chinese with English abstract) |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()