



Biodiv Sci ›› 2019, Vol. 27 ›› Issue (11): 1205-1220. DOI: 10.17520/biods.2019316
• Original Papers • Previous Articles Next Articles
Siqi Liang1,2,Xianchun Zhang1,Ran Wei1,*( )
)
Received:2019-10-09
Accepted:2019-12-17
Online:2019-11-20
Published:2020-01-17
Contact:
Wei Ran
Siqi Liang, Xianchun Zhang, Ran Wei. Integrative taxonomy resolved species delimitation in a fern complex: A case study of the Asplenium coenobiale complex[J]. Biodiv Sci, 2019, 27(11): 1205-1220.

Fig. 1 Variation of the frond division within the Asplenium coenobiale complex (Scale bar = 2 cm). (A) Habit of A. pulcherrimum (8573), bipinnate-pinnatisect; (B) Lamina of A. pulcherrimum (9525), bipinnate to tripinnate-pinnatisect; (C) Habit of A. coenobiale (9439-1), bipinnate-pinnatifid; (D) Frond of A. coenobiale (9524-6), bipinnate-pinnatisect; (E) Habit of A. coenobiale (9524-10), bipinnate to tripinnate-pinnatifid; (F) Basal pinnae A. pulcherrimum (9443), tripinnate to quadripinnatisect; (G) Basal pinnae A. pulcherrimum (9524-4), tripinnate to quadripinnatisect; (H) Lamina of A. coenobiale (9524-17), bipinnate to tripinnate-pinnatifid; (I) Basal pinnae A. coenobiale (9443), tripinnate-pinnatisect; (J) Lamina of A. coenobiale (9524-16), bipinnate.
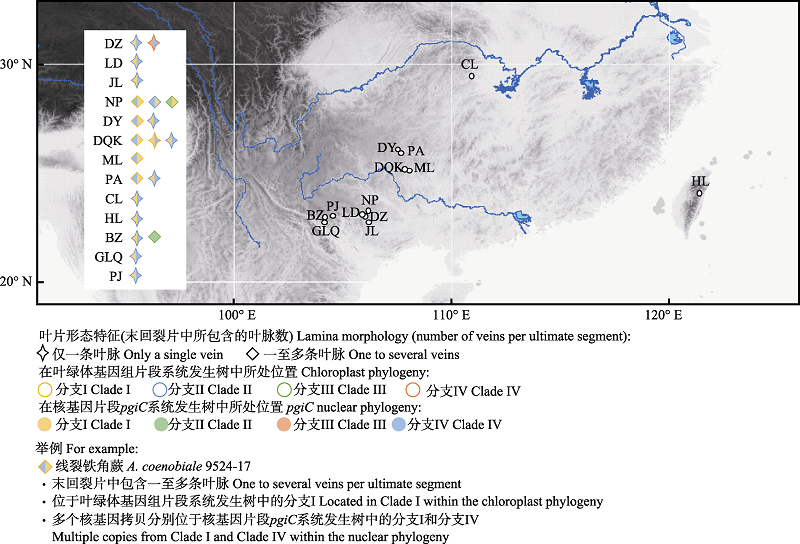
Fig. 2 Map of localities of the examined accessions of the Aspelnium coenobiale complex. DZ, LD, JL, NP, DY, DQK, ML, PA, CL, HL, BZ, GLQ, PJ are the same as Table 1.
| 片段 Fragment | 引物 Primer | 引物序列(5′-3′) Primer sequence (5′-3′) | 最佳碱基替代模型 Best substitution model | 长度 Length (bp) | 参考文献 Reference |
|---|---|---|---|---|---|
| trnL-trnF | Fern-1 | GGCAGCCCCCARATTCAGGGRAACC | K81UF+G | 693 | Trewick et al, 2002 |
| f | ATTTGAACTGGTGACACGAG | Taberlet et al, 1991 | |||
| rbcL | 1F | ATGTCACCACAAACAGA(G/A)ACTAAAGC | GTR+I+G | 1,197 | Gastony & Rollo, 1995 |
| 1351R | CTTCACAAGCAGCAGCTAGTTCAGGACTCC | Gastony & Rollo, 1995 | |||
| rpl32-trnP | 112F | TCCATCTTAACCGGTCGTCGTTCA | TVM+G | 644 | Liang et al, 2019 |
| 858R | AGTTTGGTAGCGCGTCATCT | Liang et al, 2019 | |||
| pgiC | 14F | GTGCTTCTGGGTCTTTTGAGTG | HKY+F+G4 | 648 | Ishikawa et al, 2002 |
| 16R | GTTGTCCATTAGTTCCAGGTTCCCC | Ishikawa et al, 2002 |
Table 2 Information of molecular markers in this study
| 片段 Fragment | 引物 Primer | 引物序列(5′-3′) Primer sequence (5′-3′) | 最佳碱基替代模型 Best substitution model | 长度 Length (bp) | 参考文献 Reference |
|---|---|---|---|---|---|
| trnL-trnF | Fern-1 | GGCAGCCCCCARATTCAGGGRAACC | K81UF+G | 693 | Trewick et al, 2002 |
| f | ATTTGAACTGGTGACACGAG | Taberlet et al, 1991 | |||
| rbcL | 1F | ATGTCACCACAAACAGA(G/A)ACTAAAGC | GTR+I+G | 1,197 | Gastony & Rollo, 1995 |
| 1351R | CTTCACAAGCAGCAGCTAGTTCAGGACTCC | Gastony & Rollo, 1995 | |||
| rpl32-trnP | 112F | TCCATCTTAACCGGTCGTCGTTCA | TVM+G | 644 | Liang et al, 2019 |
| 858R | AGTTTGGTAGCGCGTCATCT | Liang et al, 2019 | |||
| pgiC | 14F | GTGCTTCTGGGTCTTTTGAGTG | HKY+F+G4 | 648 | Ishikawa et al, 2002 |
| 16R | GTTGTCCATTAGTTCCAGGTTCCCC | Ishikawa et al, 2002 |
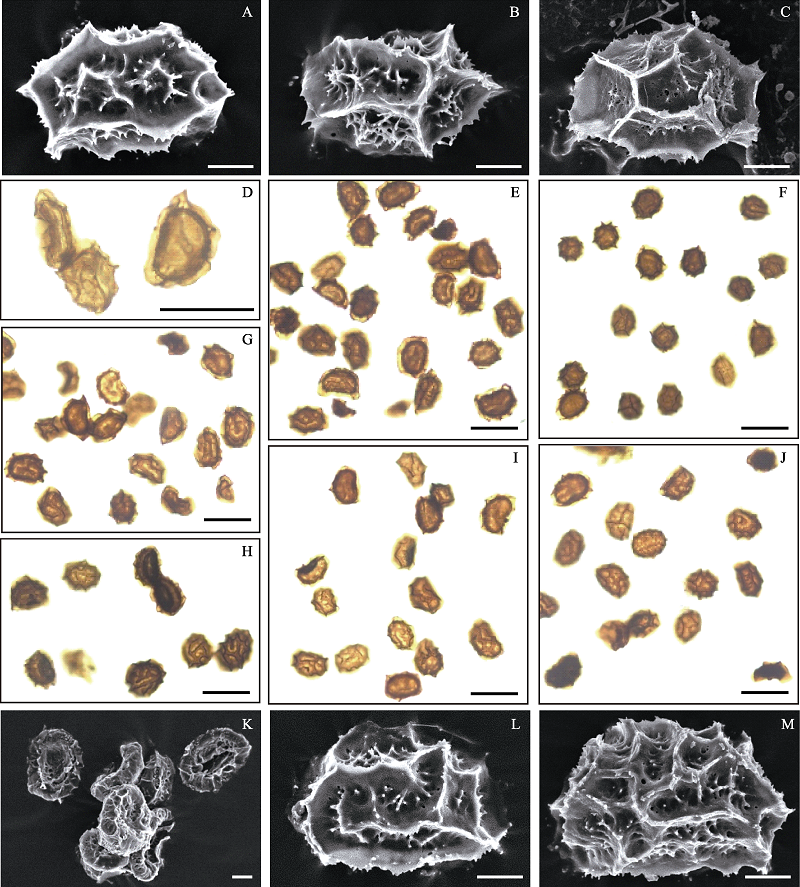
Fig. 3 Spores of the Asplenium coenobiale complex. (A) A. aff. coenobiale (8214), a normal spore, perispore lophate (cristate-alate); (B) A. aff. coenobiale (9439-2), a normal spore, perispore lophate (cristate-alate); (C) A. cornutissimum (9444), a normal spore, perispore alate; (D) A. aff. coenobiale (8214), a normal spore (kidney-shaped, well-filled) and abortive spores (shrivelled); (E) A. aff. coenobiale (9439-2), normal spores and several misshapen, blackened spores; (F) A. cornutissimum (9444), normal spores, and the size of which is relatively small compared with other accessions’ spores; (G) A. coenobiale (9524-10), normal spores and several misshapen, shrivelled spores; (H) A. coenobiale (9524-16), normal spores and several misshapen spores; (I) A. pulcherrimum (9524-1), normal spores and several blackened spores; (J) A. pulcherrimum (9443), normal spores and several misshapen, blackened spores; (K) A. coenobiale (8057), shrivelled, abortive spores; (L) A. coenobiale (9524-17), a normal spore, perispore lophate (cristate-alate); (M) A. pulcherrimum (8059B), a normal spore, perispore lophate (cristate–alate). Scale bars in A–Cand K–M are 10 μm; scale bars in D–J are 50 μm.
| 凭证标本 Voucher specimen | 核DNA含量 Nuclear DNA content (pg) | 孢子外壁长度 Length of exospore (μm) | 推定倍性 Inferred ploidy level | 孢子囊内孢子数(统计的孢子囊个数) Spore number per sporangium (Number of checked sporangia) | |
|---|---|---|---|---|---|
| 2C | 1Cx | ||||
| A. cornutissimum 9444 | 11.3 ± 0.5 | 5.7 ± 0.2 | (22-) 25-27-29 (-30) | 2x | 64 (10) |
| A. pulcherrimum 9443 | 15.8 ± 0.1 | 4.0 ± 0.0 | (29-) 32-35-37 (-39) | 4x | 64 (4) |
| A. pulcherrimum 9525 | 16.1 ± 0.5 | 4.0 ± 0.1 | (26-) 30-33-36 (-37) | 4x | 64 (2) |
| A. pulcherrimum 8059B | 16.7 | 4.2 | - | 4x | 64 (2) |
| A. pulcherrimum 9524-1 | 17.3 ± 0.2 | 4.3 ± 0.0 | (28-) 31-34-38 (-41) | 4x | 64 (6) |
| A. coenobiale 9524-13 | 17.4 ± 0.0 | 4.3 ± 0.0 | - | 4x | 64 (6) |
| A. pulcherrimum 9440 | 17.5 ± 0.2 | 4.4 ± 0.1 | - | 4x | - |
| A. coenobiale 9524-10 | 17.6 ± 0.4 | 4.4 ± 0.1 | (28-) 29-33-36 (-41) | 4x | 64 (4) |
| A. coenobiale 8059A | 17.9 | 4.5 | - | 4x | - |
| A. coenobiale 8057 | 18.3 ± 0.1 | 4.6 ± 0.0 | - | 4x | 64 (4) |
| A. coenobiale 9505 | 18.3 ± 0.1 | 4.6 ± 0.0 | - | 4x | 64 (3) |
| A. coenobiale 9524-17 | 18.3 ± 0.2 | 4.6 ± 0.0 | - | 4x | 64 (2) |
| A. coenobiale 9524-16 | 18.3 ± 0.3 | 4.6 ± 0.1 | (30-) 31-33-35 (-37) | 4x | 64 (4) |
| A. coenobiale 9439-1 | 18.7 ± 0.2 | 4.7 ± 0.1 | - | 4x | 64 (2) |
| A. coenobiale 9524-4 | 18.7 ± 0.4 | 4.7 ± 0.1 | - | 4x | 64 (4) |
| A. aff. coenobiale 9439-2 | 21.2 ± 0.2 | 5.3 ± 0.1 | (28-) 30-33-35 (-39) | 4x | 64 (1) |
| A. aff. coenobiale 8214 | 21.2 ± 0.2 | 5.3 ± 0.1 | (29-) 31-33-35 (-36) | 4x | 64 (8) |
| A. pulcherrimum 8245 | - | - | (27-) 30-32-35 (-39) | 4x | 64 (3) |
| A. pulcherrimum 2016002 | - | - | - | - | 64 (4) |
| A. coenobiale 2016017 | - | - | - | - | 64 (2) |
Table 3 Results from cytological and palynological examinations of the Asplenium coenobiale complex. -, Data missing.
| 凭证标本 Voucher specimen | 核DNA含量 Nuclear DNA content (pg) | 孢子外壁长度 Length of exospore (μm) | 推定倍性 Inferred ploidy level | 孢子囊内孢子数(统计的孢子囊个数) Spore number per sporangium (Number of checked sporangia) | |
|---|---|---|---|---|---|
| 2C | 1Cx | ||||
| A. cornutissimum 9444 | 11.3 ± 0.5 | 5.7 ± 0.2 | (22-) 25-27-29 (-30) | 2x | 64 (10) |
| A. pulcherrimum 9443 | 15.8 ± 0.1 | 4.0 ± 0.0 | (29-) 32-35-37 (-39) | 4x | 64 (4) |
| A. pulcherrimum 9525 | 16.1 ± 0.5 | 4.0 ± 0.1 | (26-) 30-33-36 (-37) | 4x | 64 (2) |
| A. pulcherrimum 8059B | 16.7 | 4.2 | - | 4x | 64 (2) |
| A. pulcherrimum 9524-1 | 17.3 ± 0.2 | 4.3 ± 0.0 | (28-) 31-34-38 (-41) | 4x | 64 (6) |
| A. coenobiale 9524-13 | 17.4 ± 0.0 | 4.3 ± 0.0 | - | 4x | 64 (6) |
| A. pulcherrimum 9440 | 17.5 ± 0.2 | 4.4 ± 0.1 | - | 4x | - |
| A. coenobiale 9524-10 | 17.6 ± 0.4 | 4.4 ± 0.1 | (28-) 29-33-36 (-41) | 4x | 64 (4) |
| A. coenobiale 8059A | 17.9 | 4.5 | - | 4x | - |
| A. coenobiale 8057 | 18.3 ± 0.1 | 4.6 ± 0.0 | - | 4x | 64 (4) |
| A. coenobiale 9505 | 18.3 ± 0.1 | 4.6 ± 0.0 | - | 4x | 64 (3) |
| A. coenobiale 9524-17 | 18.3 ± 0.2 | 4.6 ± 0.0 | - | 4x | 64 (2) |
| A. coenobiale 9524-16 | 18.3 ± 0.3 | 4.6 ± 0.1 | (30-) 31-33-35 (-37) | 4x | 64 (4) |
| A. coenobiale 9439-1 | 18.7 ± 0.2 | 4.7 ± 0.1 | - | 4x | 64 (2) |
| A. coenobiale 9524-4 | 18.7 ± 0.4 | 4.7 ± 0.1 | - | 4x | 64 (4) |
| A. aff. coenobiale 9439-2 | 21.2 ± 0.2 | 5.3 ± 0.1 | (28-) 30-33-35 (-39) | 4x | 64 (1) |
| A. aff. coenobiale 8214 | 21.2 ± 0.2 | 5.3 ± 0.1 | (29-) 31-33-35 (-36) | 4x | 64 (8) |
| A. pulcherrimum 8245 | - | - | (27-) 30-32-35 (-39) | 4x | 64 (3) |
| A. pulcherrimum 2016002 | - | - | - | - | 64 (4) |
| A. coenobiale 2016017 | - | - | - | - | 64 (2) |
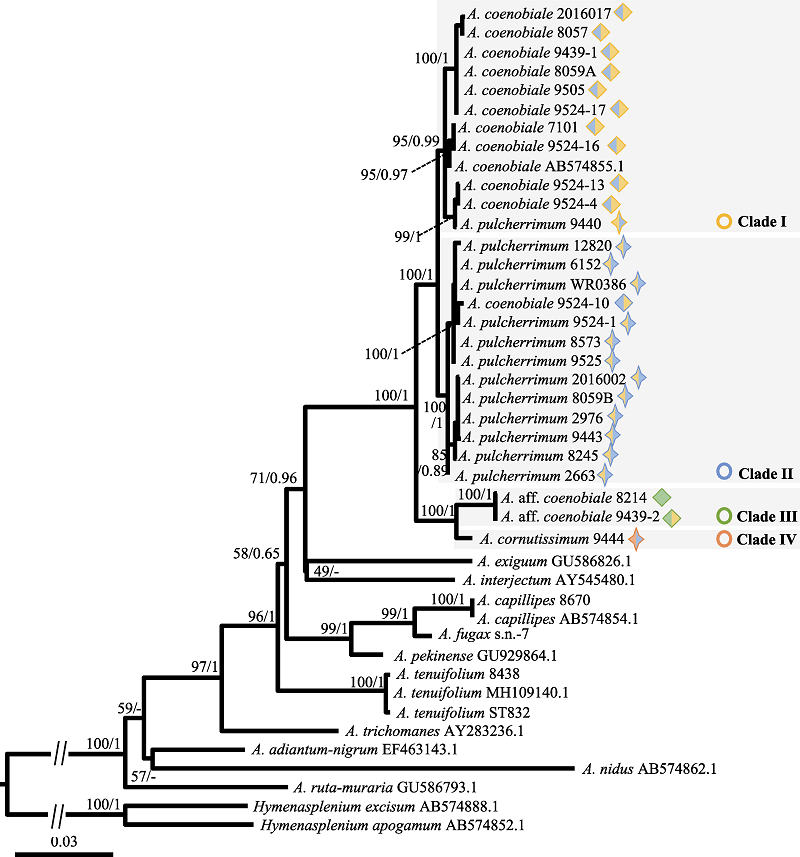
Fig. 4 The phylogenetic tree inferred from the maximum likelihood (ML) analysis based on three chloroplast DNA sequences. Numbers above the branches indicate bootstrap values or posterior probability by ML (BSML)/Bayesian inference (PPBI). Meanings of symbols are the same as Fig. 2.
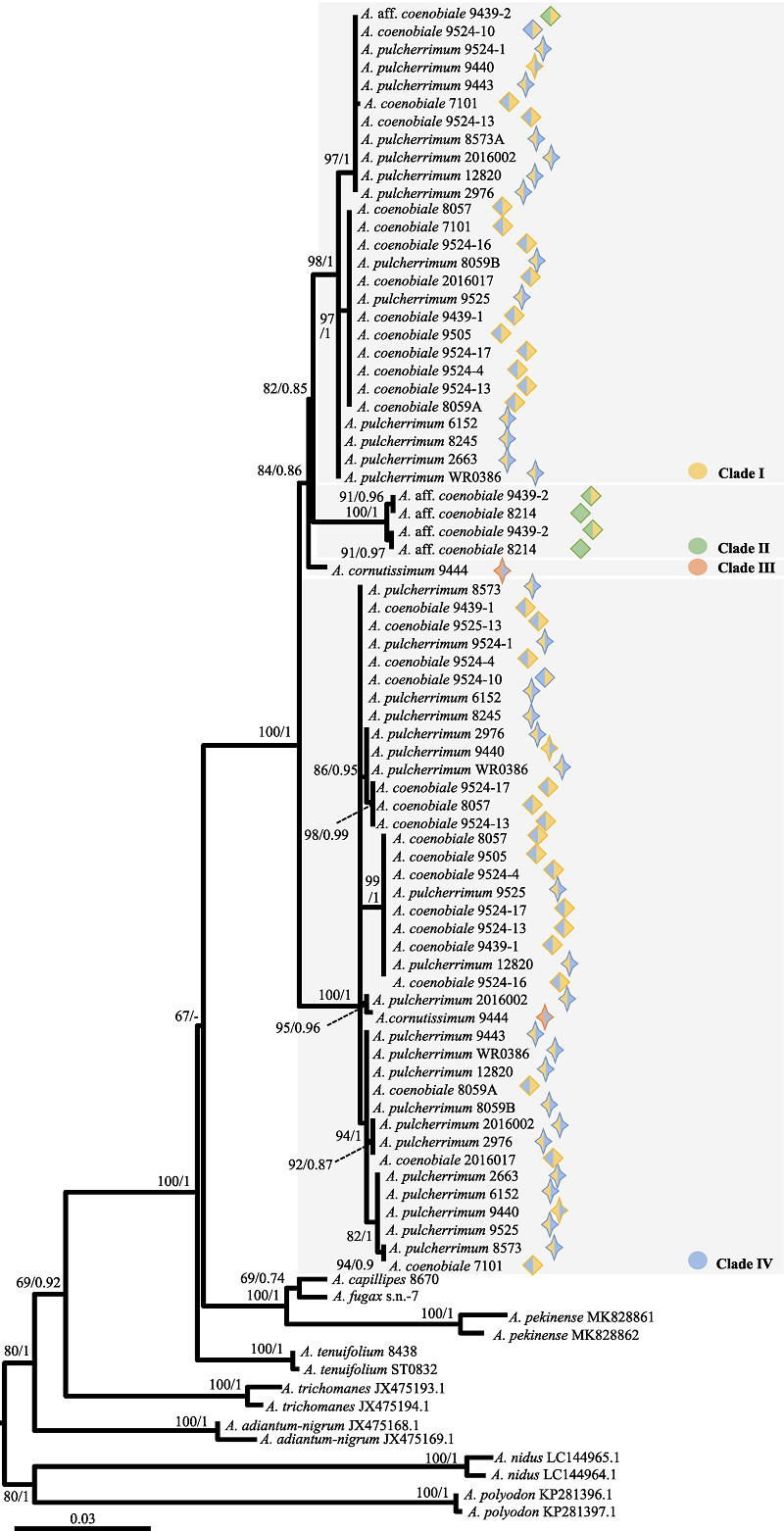
Fig. 5 The phylogenetic tree inferred from the maximum likelihood analysis based on pgiC sequence. Numbers above the branches indicate bootstrap values or posterior probability by ML (BSML)/Bayesian inference (PPBI). Meanings of symbols are the same as Fig. 2.

Fig. 6 A. maguanense sp. nov. (8214). (A) Habit; (B) Abaxial side of lamina; (C) Adaxial side of lamina; (D) Adaxial side of basal pinna; (E) Abaxial side of middle pinna; (F) Sorus; (G) Abaxial side of ultimate segment, apex with teeth and hydathodes; (H) Scales. A. aff. coenobiale (9439-2), a suspected hybrid shared half genome with A. maguanense: (I) Habitat; (J) Habit; (K) Abaxial side of ultimate segment. Scale bars in A-C and J-K are 2 cm; scale bars in D-H are 1 mm.
| 1 | Akaike H ( 1973) Information theory as an extension of the maximum likelihood principle. In: Second International Symposium on Information Theory (eds Petrov BN, Csaki F), pp. 267-281. Akademiai Kiado, Budapest. |
| 2 | Barrington DS, Paris CA, Ranker TA ( 1986) Systematic inferences from spore and stomate size in the ferns. American Fern Journal, 76, 149-159. |
| 3 | Chang YF ( 2017) Polyploidy and the formation of species diversity in Aspleniaceae. Biodiversity Science, 25, 621-626. (in Chinese with English abstract) |
| [ 常艳芬 ( 2017) 铁角蕨科的多倍化与物种多样性形成. 生物多样性, 25, 612-626.] | |
| 4 | Chang YF, Ebihara A, Lu SG, Liu HM, Schneider H ( 2018) Integrated taxonomy of the Asplenium normale complex (Aspleniaceae) in China and adjacent areas. Journal of Plant Research, 131, 573-587. |
| 5 | DeVol CE ( 1975) Aspleniaceae. In: Flora of Taiwan, Vol. 1 (ed. Editorial Committee of Flora of Taiwan). Epoch Publishing, Taipei. |
| 6 | Díez CM, Gaut BS, Meca E, Scheinvar E, Montes-Hernandez S, Eguiarte LE, Tenaillon MI ( 2013) Genome size variation in wild and cultivated maize along altitudinal gradients. New Phytologist, 199, 264-276. |
| 7 | Dyer RJ, Savolainen V, Schneider H ( 2012) Apomixis and reticulate evolution in the Asplenium monanthes fern complex. Annals of Botany, 110, 1515-1529. |
| 8 | Ebihara A, Nitta JH, Ito M ( 2010) Molecular species identification with rich floristic sampling: DNA barcoding the pteridophyte flora of Japan. PLoS ONE, 5, e15136. |
| 9 | Gastony GJ, Rollo DR ( 1995) Phylogeny and generic circumscription of cheilanthoid ferns (Pteridaceae: Cheilanthoideae) inferred from rbcL nucleotide sequences. American Fern Journal, 85, 341-360. |
| 10 | Hall TA ( 1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symposium Series, 41, 95-98. |
| 11 | Ishikawa H, Watano Y, Kano K, Ito M, Kurita S ( 2002) Development of primer sets for PCR amplification of the pgiC gene in ferns. Journal of Plant Research, 115, 65-70. |
| 12 | Jiang RH, Zhang XC, Liu Y ( 2011) Asplenium cornutissimum (Aspleniaceae), a new species from karst caves in Guangxi, China. Brittonia, 63, 83-86. |
| 13 | Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS ( 2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587-589. |
| 14 | Kumar S, Stecher G, Tamura K ( 2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874. |
| 15 | Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B ( 2016) PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34, 772-773. |
| 16 | Leitch IJ, Bennett MD ( 2004) Genome downsizing in polyploid plants. Biological Journal of the Linnean Society, 82, 651-663. |
| 17 | Li JZ ( 2004) Aspleniaceae. In: Flora of Hunan, Vol. 1 (eds Li JZ,Chen SM, Lin QZ).Hunan Science & Technology Press, Changsha. (in Chinese) |
| [ 李建宗 ( 2004) 铁角蕨科. 见: 湖南植物志第一卷 (李建宗, 陈三茂, 林亲众编). 湖南科学技术出版社, 长沙.] | |
| 18 | Liang SQ, Viane RLL, Zhang XC, Wei R ( 2019) Exploring the reticulate evolution in the Asplenium pekinense complex and the A. varians complex (Aspleniaceae). Journal of Systematics and Evolution, doi: 10.1111/jse.12530. |
| 19 | Lin YX, Sleep A ( 1989) A cytogenetic study of two Asplenium species from Eastern Asia: A. sarelii and A. pekinense (Aspleniaceae: Pteridophyta). In: Proceedings of the International Symposium on Systematic Pteridology (eds Shing KH, Kramer KU), pp. 111-127. China Science and Technology Press, Beijing. |
| 20 | Lin YX, Viane R ( 2013) Aspleniaceae. In: Flora of China, Vol.2-3(eds Wu ZY, Raven P, Hong DY). Science Press, Beijing & Missouri Botanical Garden Press, St. Louis. |
| 21 | Lovis JD ( 1978) Evolutionary patterns and processes in ferns. Advances in Botanical Research, 4, 229-415. |
| 22 | Lovis JD, Reichstein T ( 1964) A diploid form of Asplenium ruta-muraria. British Fern Gazette, 9, 141-146. |
| 23 | Mallet J ( 2007) Hybrid speciation. Nature, 446, 279-283. |
| 24 | Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ ( 2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268-274. |
| 25 | Ohlsen DJ, Perrie LR, Shepherd LD, BrownseyPJ, Bayly MJ ( 2015) Investigation of species boundaries and relationships in the Asplenium paleaceum complex (Aspleniaceae) using AFLP fingerprinting and chloroplast and nuclear DNA sequences. Australian Systematic Botany, 27, 378-394. |
| 26 | Otto F ( 1990) DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Methods in Cell Biology (eds Crissman HA, Dsrzynkiewicz Z), pp. 105-110. Academic Press, New York. |
| 27 | PPG I ( 2016) A community based classification of ferns and lycophytes. Journal of Systematics and Evolution, 54, 563-603. |
| 28 | Rieseberg LH, Willis JH ( 2007) Plant speciation. Science, 317, 910-914. |
| 29 | Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP ( 2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539-542. |
| 30 | Schneider H, Liu HM, Chang YF, Ohlsen D, Perrie LR, Shepherd L, Kessler M, Karger D, Hennequin S, Marquardt J, Russell S, Ansell S, Lu NT, Kamau P, Regalado JL, Heinrichs L, Ebihara J, Smith A, Gibby M ( 2017) Neo- and Palaeopolyploidy contribute to the species diversity of Asplenium—The most species rich genus of ferns. Journal of Systematics and Evolution, 55, 353-364. |
| 31 | Shang H, Yan YH ( 2017) Natural hybridization and biodiversity conservation. Biodiversity Science, 25, 683-688. (in Chinese with English abstract) |
| [ 商辉, 严岳鸿 ( 2017) 自然杂交与生物多样性保护. 生物多样性, 25, 683-688.] | |
| 32 | Sigel EM ( 2016) Genetic and genomic aspects of hybridization in ferns. Journal of Systematics and Evolution, 54, 638-655. |
| 33 | Soltis DE, Visger CJ, Soltis PS ( 2014) The polyploidy revolution then…and now: Stebbins revisited. American Journal of Botany, 101, 1057-1078. |
| 34 | Soltis PS, Marchant DB, van de Peer Y, Soltis DE ( 2015) Polyploidy and genome evolution in plants. Current Opinion in Genetics & Development, 35, 119-125. |
| 35 | Soltis PS, Soltis DE ( 2009) The role of hybridization in plant speciation. Annual Review of Plant Biology, 60, 561-588. |
| 36 | Taberlet P, Gielly L, Pautou G, Bouvet J ( 1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology, 17, 1105-1109. |
| 37 | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG ( 1997) The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. |
| 38 | Trewick SA, Morgan-Richards M, Russell SJ, Henderson S, Rumsey FJ, Pinter I, Barrett JA, Gibby M, Vogel JC ( 2002) Polyploidy, phylogeography and Pleistocene refugia of the rockfern Asplenium ceterach: Evidence from chloroplast DNA. Molecular Ecology, 11, 2003-2012. |
| 39 | Viane RLL, Reichstein T ( 2003) Notes on new or interesting Asplenium species from western Asia, including comments on Ching & Wu (1985), and Fraser-Jenkins (1992) Reliquiae Reichsteinianae 1. In: Pteridology in the New Millennium (eds Chandra S, Srivastava M), pp. 73-105. Springer, Dordrecht. |
| 40 | Vogel JC, Russell SJ, Rumsey FJ, Barrett JA, Gibby M ( 1998) Evidence for maternal transmission of chloroplast DNA in the genus Asplenium (Aspleniaceae, Pteridophyta). Botanica Acta, 111, 247-249. |
| 41 | Wagner WH ( 1954) Reticulate evolution in the Appalachian Asplenium. Evolution, 8, 103-118. |
| 42 | Wang PS, Wang XY ( 2001) Aspleniaceae. In: Pteridophyte Flora of Guizhou (eds Wang PS, Wang XY). Guizhou Science & Technology Publishing House, Guiyang. (in Chinese) |
| [ 王培善, 王筱英 ( 2001) 铁角蕨科. 见:贵州蕨类植物志(王培善, 王筱英编). 贵州科技出版社, 贵阳.] | |
| 43 | Wang YG ( 2017) Natural hybridization and speciation. Biodiversity Science, 25, 565-576. (in Chinese with English abstract) |
| [ 王玉国 ( 2017) 自然杂交与物种形成. 生物多样性, 25, 565-576.] | |
| 44 | Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH ( 2009) The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA, 106, 13875-13879. |
| 45 | Wu ZH ( 1999) Aspleniaceae. In: Flora Reipublicae Popularis Sinicae (ed. Editorial Committee of Flora Reipublicae Popularis Sinicae, Chinese Academy of Sciences), Tomus 4(2).Science Press, Beijing. (in Chinese) |
| [ 吴兆洪 ( 1999) 铁角蕨科. 见:中国植物志, 第4卷, 第2分册 (中国科学院中国植物志编辑委员会编). 科学出版社, 北京.] | |
| 46 | Wu ZH ( 2006) Aspleniaceae. In: Flora of Guangdong, Vol. VII (ed. Wu TL). Guangdong Science and Technology Press, Guangzhou. (in Chinese) |
| [ 吴兆洪 ( 2006) 铁角蕨科. 见:广东植物志, 第7卷 (吴德邻编).广东科技出版社, 广州.] | |
| 47 | Xu KW, Zhang L, Rothfels CJ, Smith AR, Viane R, Lorence D, Wood KR, Chen CW, Knapp R, Zhou L, Lu NT, Zhou XM, Wei HJ, Fan Q, Chen SF, Cicuzza D, Gao XF, Liao WB, Zhang LB ( 2019) A global plastid phylogeny of the fern genus Asplenium (Aspleniaceae). Cladistics , doi: 10.1111/cla. |
| 12384. | |
| 48 | Yakimowski SB, Rieseberg LH ( 2014) The role of homoploid hybridization in evolution: A century of studies synthesizing genetics and ecology. American Journal of Botany, 101, 1247-1258. |
| 49 | Zhang D, Gao FL, Li WX, Jakovlić I, Zou H, Zhang J, Wang GT ( 2018) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. bioRxiv , doi: 10.1101/489088. |
| 50 | Zhang GF ( 2006) Aspleniaceae. In: Flora Yunnanica, Tomus 20 (Pterisophyta) (ed. Zhu WM). Science Press, Beijing. (in Chinese) |
| [ 张光飞 (2006) 铁角蕨科. 见:云南植物志, 第二十卷 (蕨类植物) (朱维明编). 科学出版社, 北京.] |
| [1] | Chenyang Xue, Yufeng Xu, Bo Qu. Comparison of morphology, photosynthesis, and growth among Xanthium strumarium, X. sibiricum and their hybrid under different nitrogen levels [J]. Biodiv Sci, 2018, 26(6): 554-563. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()