



Biodiv Sci ›› 2017, Vol. 25 ›› Issue (12): 1339-1349. DOI: 10.17520/biods.2017265 cstr: 32101.14.biods.2017265
• Review • Previous Articles Next Articles
Junwei Ye, Yongge Yuan, Li Cai, Xiaojuan Wang*( )
)
Received:2017-10-12
Accepted:2017-12-04
Online:2017-12-20
Published:2017-12-10
Contact:
Wang Xiaojuan
Junwei Ye, Yongge Yuan, Li Cai, Xiaojuan Wang. Research progress of phylogeographic studies of plant species in temperate coniferous and broadleaf mixed forests in Northeastern China[J]. Biodiv Sci, 2017, 25(12): 1339-1349.
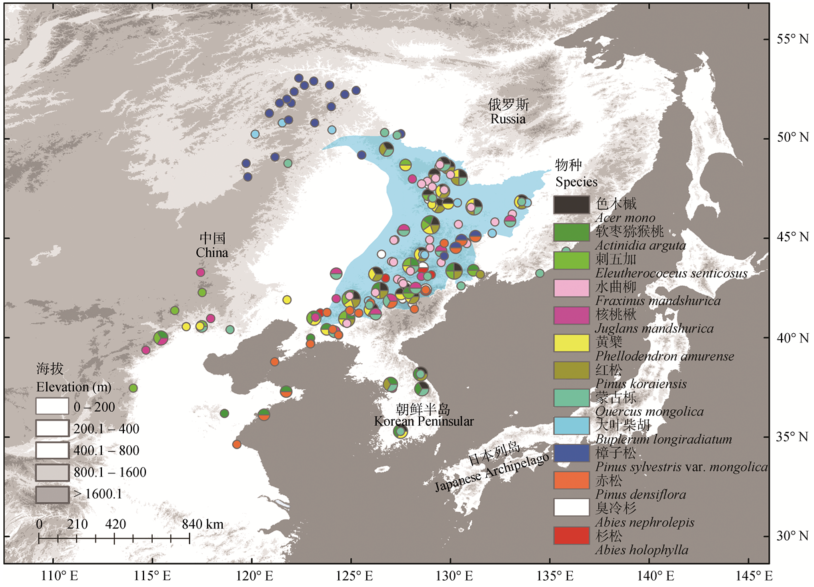
Fig. 1 Distribution of sampled locations in phylogeographic studies (Table 1) of temperate coniferous and broadleaf mixed forest (blue region) plants in Northeast China
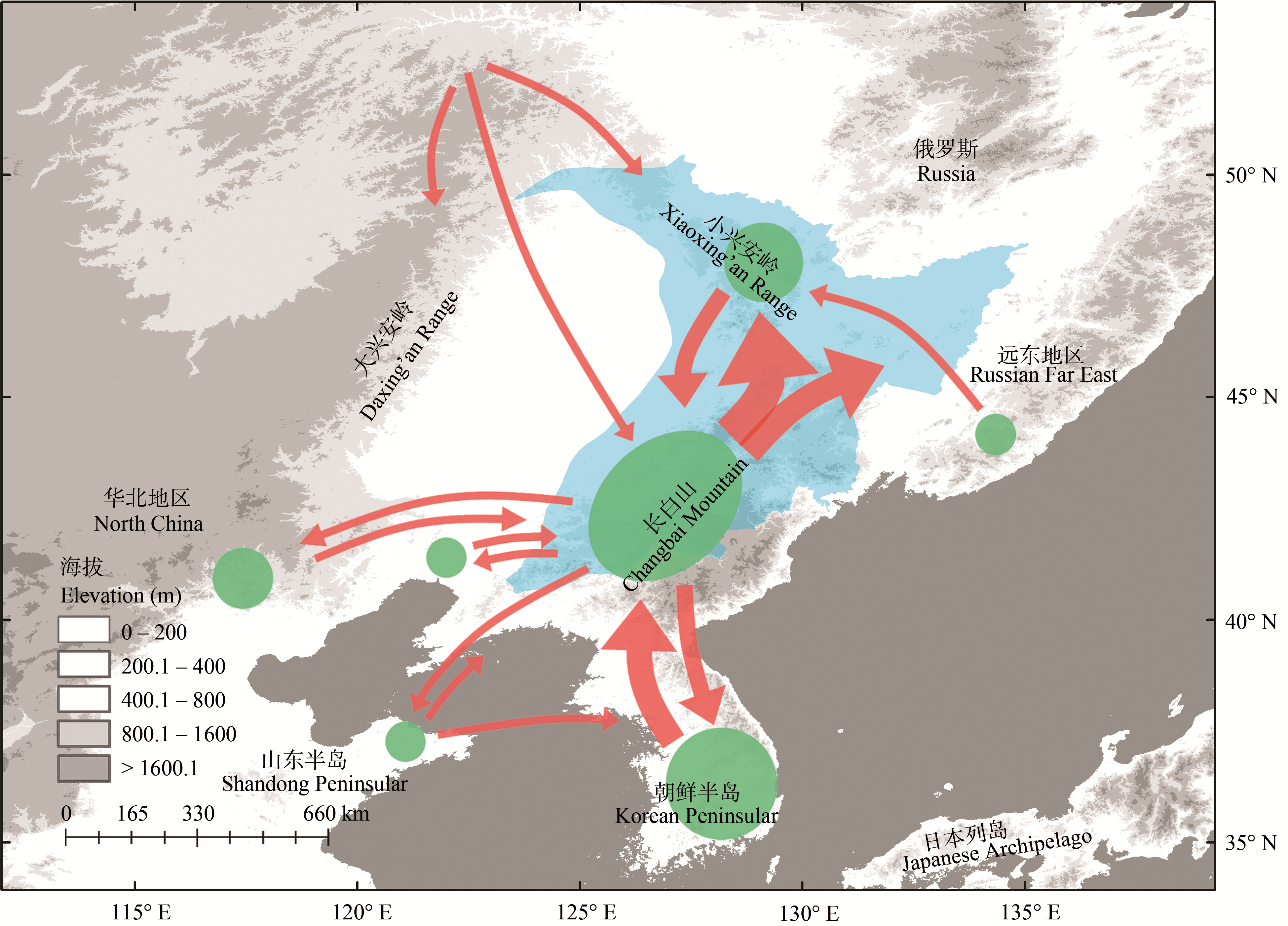
Fig. 2 Refugia and expansion routes of temperate coniferous and broadleaf mixed forest (blue region) plants in Northeast China. Refugia area size and expansion route line thickness are proportional to the number of related inference in previous studies (Table 1).
| [1] | Aizawa M, Yoshimaru H, Saito H, Katsuki T, Kawahara T, Kitamura K, Shi F, Kaji M (2007) Phylogeography of a northeast Asian spruce, Picea jezoensis, inferred from genetic variation observed in organelle DNA markers. Molecular Ecology, 16, 3393-3405. |
| [2] | Ashcroft MB (2010) Identifying refugia from climate change. Journal of Biogeography, 37, 1407-1413. |
| [3] | Avise JC (2000) Phylogeography: The History and Formation of Species. Harvard University Press, Cambridge, MA. |
| [4] | Axelrod DI, Al-Shehbaz I, Raven PH (1996) History of the modern flora of China. In: Floristic Characteristics and Diversity of East Asian Plants (eds Zhang A, Wu SG). China Higher Education Press/Springer-Verlag Press, Beijing. |
| [5] | Bai WN, Liao WJ, Zhang DY (2010) Nuclear and chloroplast DNA phylogeography reveal two refuge areas with asymmetrical gene flow in a temperate walnut tree from East Asia. New Phytologist, 188, 892-901. |
| [6] | Bai WN, Zhang DY (2014) Current status and future direction in plant phylogeography. Chinese Bulletin of Life Sciences, 26, 125-137. (in Chinese with English abstract) |
| [白伟宁, 张大勇 (2014) 植物亲缘地理学的研究现状与发展趋势. 生命科学, 26, 125-137.] | |
| [7] | Bai WN, Wang WT, Zhang DY (2016) Phylogeographic breaks within Asian butternuts indicate the existence of a phytogeo¬graphic divide in East Asia. New Phytologist, 209, 1757-1772. |
| [8] | Bao L, Ayijiamali K, Bai WN, Chen RZ, Wang TM, Wang HF, Ge JP (2015) Contributions of multiple refugia during the last glacial period to current mainland populations of Korean pine (Pinus koraiensis). Scientific Reports, 5, 18608. |
| [9] | Cartens BC, Brunsfeld SJ, Demboski JR, Good JM, Sullivan J (2005) Investigating the evolutionary history of the pacific northwest mesic forest ecosystem: hypothesis testing wit¬hin a comparative phylogeographic framework. Evolution, 59, 1639-1652. |
| [10] | Cavender-Bares J, Kozak KH, Fine PV, Kembel SW (2009) The merging of community ecology and phylogenetic biology. Ecology Letters, 12, 693-715. |
| [11] | Chen KM, Abbott RJ, Milne RI, Tian XM, Liu JQ (2008) Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a do¬minant species of coniferous forest in northern China. Mol¬ecular Ecology, 17, 4276-4288. |
| [12] | Clark PU, Mccabe AM (2009) The Last Glacial Maximum. Science, 325, 710-714. |
| [13] | Excoffier L, Ray N (2008) Surfing during population expansions promotes genetic revolutions and structuration. Trends in Ecology & Evolution, 23, 347-351. |
| [14] | Gavin DG, Fitzpatrick MC, Gugger PF, Heath KD, Francisco RS, Dobrowski SZ, Hampe A, Sheng HF, Ashcroft MB, Bartlein PJ (2014) Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytologist, 204, 37-54. |
| [15] | Guo XD, Wang HF, Bao L, Bai WN, Ge JP (2014) Asymmetric pattern of climate oscillations and the origin of biodiversity. Journal of Beijing Normal University (Natural Science), 50, 622-628. (in Chinese with English abstract) |
| [郭希的, 王红芳, 鲍蕾, 白伟宁, 葛剑平 (2014) 冰期非对称气候变化模式与生物多样性的形成机制. 北京师范大学学报(自然科学版), 50, 622-628.] | |
| [16] | Guo XD, Wang HF, Bao L, Wang TM, Bai WN, Ye JW, Ge JP (2014) Evolutionary history of a widespread tree species Acer mono in East Asia. Ecology and Evolution, 4, 4332-4345. |
| [17] | Harrison SP, Yu G, Takahara H, Prentice IC (2001) Palaeovegetation (Communications arising): diversity of temperate plants in East Asia. Nature, 413, 129-130. |
| [18] | Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society, 58, 247-276. |
| [19] | Hewitt GM (1999) Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society, 68, 87-112. |
| [20] | Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature, 405, 907-913. |
| [21] | Hickerson MJ, Carstens BC, Cavender-bares J, Crandall KA, Graham CH, Johnson JB, Rissler L, Victoriano PF, Yoder AD (2010) Phylogeography’s past, present, and future: 10 years after Avise, 2000. Molecular Phylogenetics and Evolution, 54, 291-301. |
| [22] | Hu LJ, Uchiyama K, Shen HL, Saito Y, Tsuda Y, Ide Y (2008) Nuclear DNA microsatellites reveal genetic variation but a lack of phylogeographical structure in an endangered species, Fraxinus mandshurica, across North-east China. Annals of Botany, 102, 195-205. |
| [23] | Ibrahim KM, Nichols GM, Hewitt GM (1996) Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity, 77, 282-291. |
| [24] | Jaramillo-Correa JP, Beaulieu J, Khasa DP, Bousquet J (2009) Inferring the past from the present phylogeographic structure of North American forest trees: seeing the forest for the genes. Canadian Journal of Forest Research, 39, 286-307. |
| [25] | Jiang ZY, Peng YL, Hu XX, Zhou YF, Liu JQ (2011) Cytoplasmic DNA variation in and genetic delimitation of Abies nephrolepis and Abies holophylla in northeastern China. Canadian Journal of Forest Research, 41, 1555-1561. |
| [26] | Keppel G, Niel KPV, Wardell-Johnson GW, Yates CJ, Byrne M, Mucina L, Schut AGT (2012) Refugia: identifying and understanding safe havens for biodiversity under climate change. Global Ecology and Biogeography, 21, 393-404. |
| [27] | Lafontaine GD, Ducousso A, Lefèvre S, Magnanou E, Petit RJ (2013) Stronger spatial genetic structure in recolonized areas than in refugia in the European beech. Molecular Ecology, 22, 4397-4412. |
| [28] | Lapola DM, Oyama MD, Nobre CA, Sampaio G (2008) A new world natural vegetation map for global change studies. Anais Da Academia Brasileira De Ciencias, 80, 397-408. |
| [29] | Liu CP, Tsuda Y, Shen HL, Hu LJ, Saito Y, Ide Y (2014) Genetic structure and hierarchical population divergence history of Acer mono var. mono in south and northeast China. PLoS ONE, 9, e87187. |
| [30] | Liu JQ, Sun YS, Ge XJ, Gao LM, Qiu YX (2012) Phylogeographic studies of plants in China: advances in the past and directions in the future. Journal of Systematics and Evolution, 50, 267-275. |
| [31] | López-Pujol J, Zhang FM, Sun HQ, Ying TS, Ge S (2011) Centres of plant endemism in China: places for survival or for speciation? Journal of Biogeography, 38, 1267-1280. |
| [32] | Ni J, Yu G, Harrison SP, Prentice IC (2010) Palaeovegetation in China during the late Quaternary: biome reconstructions based on a global scheme of plant functional types. Palaeogeography, Palaeoclimatology, Palaeoecology, 289, 44-61. |
| [33] | Nogués-Bravo D (2009) Predicting the past distribution of species climatic niches. Global Ecology and Biogeography, 18, 521-531. |
| [34] | Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends in Ecology & Evolution, 23, 564-571. |
| [35] | Qian H, Ricklefs RE (1999) A comparison of the taxonomic richness of vascular plants in China and the United States. The American Naturalist, 154, 160-181. |
| [36] | Qian H, Ricklefs RE (2000) Large-scale processes and the Asian bias in species diversity of temperate plants. Nature, 407, 180-182. |
| [37] | Qiu YX, Fu CX, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental cha¬nge in the world’s most diverse temperate flora. Molecular Phylogenetics and Evolution, 59, 225-244. |
| [38] | Ren GP, Abbott RJ, Zhou YF, Zhang LR, Peng YL, Liu JQ (2012) Genetic divergence, range expansion and possible homoploid hybrid speciation among pine species in Northeast China. Heredity, 108, 552-562. |
| [39] | Wu ZY (1980) Chinese Vegetation. Science Press, Beijing. (in Chinese) |
| [吴征镒 (1980) 中国植被. 科学出版社, 北京.] | |
| [40] | The Editorial Comittee of Vegetation Map of China, Chinese Academy of Sciences(2007) Vegetation Map of China and Its Geographic Pattern. Geological Publishing House, Beijing. (in Chinese) |
| [中国科学院中国植被图编辑委员会(2007) 中国植被及其地理格局. 地质出版社, 北京.] | |
| [41] | Wang LJ, Bao L, Wang HF, Ge JP (2014) Study on the genetic diversity of Phyllodendron amurense. China Sciencepaper, 9, 1397-1401. (in Chinese with English abstract) |
| [王丽君, 鲍蕾, 王红芳, 葛剑平 (2014) 黄檗遗传多样性研究. 中国科技论文, 9, 1397-1401.] | |
| [42] | Wang SH, Bao L, Wang TM, Wang HF, Ge JP (2016a) Contrasting genetic patterns between two coexisting Eleutherococcus species in northern China. Ecology and Evolution, 6, 3311-3324. |
| [43] | Wang WT, Xu B, Zhang DY, Bai WN (2016b) Phylogeography of postglacial range expansion in Juglans mandshurica (Juglandaceae) reveals no evidence of bottleneck, loss of genetic diversity, or isolation by distance in the leading-edge populations. Molecular Phylogenetics and Evolution, 102, 255-264. |
| [44] | Waters JM, Fraser CI, Hewitt GM (2013) Founder takes all: density-dependent processes structure biodiversity. Trends in Ecology & Evolution, 28, 78-85. |
| [45] | Widmer A, Lexer C (2001) Glacial refugia: sanctuaries for allelic richness, but not for gene diversity. Trends in Ecology & Evolution, 16, 267-269. |
| [46] | Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Aca¬demy of Sciences, USA, 84, 9054-9058. |
| [47] | Yang C, Cheng BD, Xie Y, Song WM (2017) Development stage and spatiotemporal evolution of China’s timber industry. Journal of Natural Resources, 32, 235-244. (in Chinese with English abstract) |
| [杨超, 程宝栋, 谢屹, 宋维明 (2017) 中国木材产业发展的阶段识别及时空分异特征. 自然资源学报, 32, 235-244.] | |
| [48] | Ye JW, Zhang Y, Wang XJ (2017) Phylogeographic history of broad-leaved forest plants in subtropical China. Acta Ecologica Sinica, 37, 5894-5904. (in Chinese with English abstract) |
| [叶俊伟, 张阳, 王晓娟 (2017) 中国亚热带地区阔叶林植物的谱系地理历史. 生态学报, 37, 5894-5904.] | |
| [49] | Ye JW, Jiang T, Wang HF, Wang TM, Bao L, Ge JP (2018) Repeated expansions and fragmentations linked to Pleistocene climate changes shaped the genetic structure of a wo¬ody climber, Actinidia arguta (Actinidiaceae). Botany, 2018, 96, 19-31. |
| [50] | Ying TS (2001) Species diversity and distribution pattern of seed plants in China. Biodiversity Science, 9, 393-398. (in Chinese with English abstract) |
| [应俊生 (2001) 中国种子植物物种多样性及其分布格局. 生物多样性, 9, 393-398.] | |
| [51] | Yu G, Chen X, Ni J, Cheddadi R, Guiot J, Han H, Harrison SP, Huang C, Ke M, Kong Z (2000) Palaeovegetation of China: a pollen data-based synthesis for the mid-Holocene and last glacial maximum. Journal of Biogeography, 27, 635-664. |
| [52] | Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science, 292, 686-693. |
| [53] | Zeng YF, Wang WT, Liao WJ, Wang HF, Zhang DY (2015) Multiple glacial refugia for cool-temperate deciduous trees in northern East Asia: the Mongolian oak as a case study. Molecular Ecology, 56, 584-591. |
| [54] | Zhang W, Niu YB, Yan L, Cui ZJ, Liu CC, Mu KH (2008) Late Pleistocene glaciation of the Changbai Mountains in northeastern China. Chinese Science Bulletin, 53, 2672-2684. |
| [55] | Zhao C, Wang CB, Ma XG, Liang QL, He XJ (2013) Phylogeographic analysis of a temperate-deciduous forest restricted plant (Bupleurum longiradiatum Turcz.) reveals two refuge areas in China with subsequent refugial isolation promoting speciation. Molecular Phylogenetics and Evolution, 68, 628-643. |
| [56] | Zheng BX, Xu QQ, Shen YP (2002) The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quaternary International, 97/98, 93-101. |
| [57] | Zheng YJ, Xiao XM, Guo ZW, Howard TE (2001) A county-level analysis of the spatial distribution of forest resources in China. Journal of Forest Planning, 7, 69-78. |
| [58] | Zhou YL (1997) Vegetation Geography in Northeast China. Science Press, Beijing. (in Chinese) |
| [周以良 (1997) 中国东北植被地理. 科学出版社, 北京.] |
| [1] | Jiachen Wang, Tangjun Xu, Wei Xu, Gaoji Zhang, Yijin You, Honghua Ruan, Hongyi Liu. Impact of urban landscape pattern on the genetic structure of Thereuopoda clunifera population in Nanjing, China [J]. Biodiv Sci, 2025, 33(1): 24251-. |
| [2] | Kexin Cao, Jingwen Wang, Guo Zheng, Pengfeng Wu, Yingbin Li, Shuyan Cui. Effects of precipitation regime change and nitrogen deposition on soil nematode diversity in the grassland of northern China [J]. Biodiv Sci, 2024, 32(3): 23491-. |
| [3] | Shiyi Long, Bobo Zhang, Yuchen Xia, Yangfan Fei, Yani Meng, Bingwei Lü, Yueqing Song, Pu Zheng, Taoran Guo, Jian Zhang, Shaopeng Li. Effects of diversity and temporal stability of native communities on the biomass of invasive species Solidago canadensis [J]. Biodiv Sci, 2024, 32(11): 24263-. |
| [4] | Linjun He, Wenjing Yang, Yuhao Shi, Kezhemo Ashuo, Yu Fan, Guoyan Wang, Jingji Li, Songlin Shi, Guihua Yi, Peihao Peng. Effects of plant community phylogeny and functional diversity on Ageratina adenophora invasion under fire disturbance [J]. Biodiv Sci, 2024, 32(11): 24269-. |
| [5] | Qingduo Li, Dongmei Li. Analysis for the prevalence of global bat-borne Bartonella [J]. Biodiv Sci, 2023, 31(9): 23166-. |
| [6] | Chen Feng, Jie Zhang, Hongwen Huang. Parallel situ conservation: A new plant conservation strategy to integrate in situ and ex situ conservation of plants [J]. Biodiv Sci, 2023, 31(9): 23184-. |
| [7] | Hailing Qi, Pengzhen Fan, Yuehua Wang, Jie Liu. Genetic diversity and population structure of Juglans regia from six provinces in northern China [J]. Biodiv Sci, 2023, 31(8): 23120-. |
| [8] | Yuanyuan Xiao, Wei Feng, Yangui Qiao, Yuqing Zhang, Shugao Qin. Effects of soil microbial community characteristics on soil multifunctionality in sand-fixation shrublands [J]. Biodiv Sci, 2023, 31(4): 22585-. |
| [9] | Fei Xiong, Hongyan Liu, Dongdong Zhai, Xinbin Duan, Huiwu Tian, Daqing Chen. Population genetic structure of Pelteobagrus vachelli in the upper Yangtze River based on genome re-sequencing [J]. Biodiv Sci, 2023, 31(4): 22391-. |
| [10] | Yiyue He, Yuying Liu, Fubin Zhang, Qiang Qin, Yu Zeng, Zhenyu Lü, Kun Yang. Genetic diversity and population structure of Saurogobio dabryi under cascade water conservancy projects in the Jialing River [J]. Biodiv Sci, 2023, 31(11): 23160-. |
| [11] | Weiyue Sun, Jiangping Shu, Yufeng Gu, Morigengaowa, Xiajin Du, Baodong Liu, Yuehong Yan. Conservation genomics analysis revealed the endangered mechanism of Adiantum nelumboides [J]. Biodiv Sci, 2022, 30(7): 21508-. |
| [12] | Xiaoyan Jiang, Shengjie Gao, Yan Jiang, Yun Tian, Xin Jia, Tianshan Zha. Species diversity, functional diversity, and phylogenetic diversity in plant communities at different phases of vegetation restoration in the Mu Us sandy grassland [J]. Biodiv Sci, 2022, 30(5): 21387-. |
| [13] | Togtokh Mongke, Dongyi Bai, Tugeqin Bao, Ruoyang Zhao, Tana An, Aertengqimike Tiemuqier, Baoyindeligeer Mongkejargal, Has Soyoltiin, Manglai Dugarjaviin, Haige Han. Assessment of SNPs-based genomic diversity in different populations of Eastern Asian landrace horses [J]. Biodiv Sci, 2022, 30(5): 21031-. |
| [14] | Jing Cui, Mingfang Xu, Qun Zhang, Yao Li, Xiaoshu Zeng, Sha Li. Differences in genetic diversity of Pleuronichthys cornutus in the coastal water of China and Japan based on three mitochondrial markers [J]. Biodiv Sci, 2022, 30(5): 21485-. |
| [15] | Xinyu Cai, Xiaowei Mao, Yiqiang Zhao. Methods and research progress on the origin of animal domestication [J]. Biodiv Sci, 2022, 30(4): 21457-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()