



生物多样性 ›› 2019, Vol. 27 ›› Issue (12): 1291-1297. DOI: 10.17520/biods.2019255 cstr: 32101.14.biods.2019255
收稿日期:2019-08-16
接受日期:2019-11-20
出版日期:2019-12-20
发布日期:2020-01-14
通讯作者:
张雁云
基金资助:
Jian Zhang,Lu Dong,Yanyun Zhang( )
)
Received:2019-08-16
Accepted:2019-11-20
Online:2019-12-20
Published:2020-01-14
Contact:
Zhang Yanyun
摘要:
我国提议在南极恩克斯堡岛新站址北侧3 km的阿德利企鹅(Pygoscelis adeliae)聚集繁殖地建立南极特别保护区, 对保护区边界的划分, 各国尚存争议, 尤其是对南湾(South Bay)的繁殖小种群是否具有遗传独特性, 是否应将其纳入保护区是重点关注的问题。本研究采集了恩克斯堡岛海景湾(Seaview Bay)和南湾的阿德利企鹅样品, 通过全基因组重测序和种群基因组学方法, 分析了恩克斯堡岛不同区域的种群遗传结构。发现恩克斯堡岛海景湾与南湾阿德利企鹅没有显著的遗传分化, 南湾阿德利企鹅不是独特的小种群; 海景湾高海拔区域个体与低海拔区域个体之间也没有显著的遗传差异, 推测该区域阿德利企鹅繁殖群体的分布格局与冰川堆积形成的阶地不具有显著相关性。本工作为恩克斯堡岛保护区和罗斯海新站建设提供了重要科技支撑。
张剑, 董路, 张雁云 (2019) 基于基因组SNPs的南极恩克斯堡岛阿德利企鹅繁殖种群的遗传结构. 生物多样性, 27, 1291-1297. DOI: 10.17520/biods.2019255.
Jian Zhang, Lu Dong, Yanyun Zhang (2019) Population genetic structure of Adélie penguins (Pygoscelis adeliae) from Inexpressible Island, Antarctica, using SNP markers. Biodiversity Science, 27, 1291-1297. DOI: 10.17520/biods.2019255.
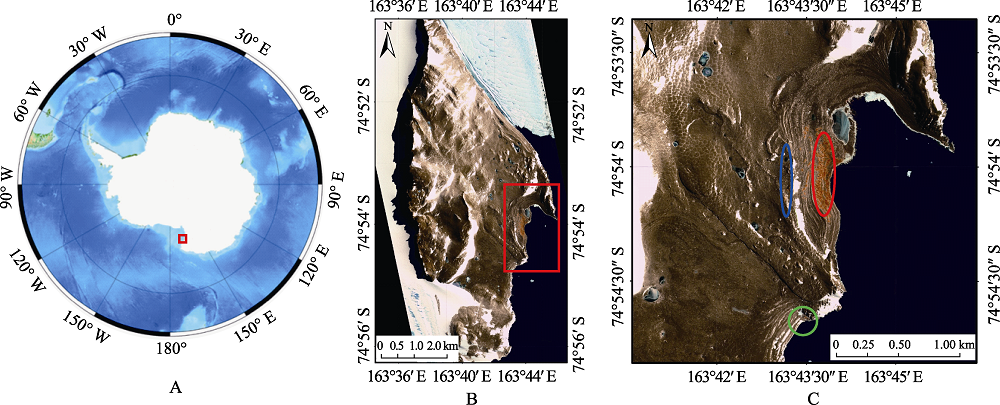
图1 阿德利企鹅采样区地图。A: 南极地图, 红框示恩克斯堡岛的位置; B: 恩克斯堡岛地图, 红框示拟建保护区的位置; C: 采样区域, 蓝色和红色圆圈分别为海景湾高海拔和低海拔采样区, 绿色圆圈为南湾采样区。
Fig. 1 Sampling sites of Adélie penguins (Pygoscelis adeliae) in Inexpressible Island. (A) Map of Antarctic (Red rectangle: Inexpressible Island). (B) Map of Inexpressible Island (Red rectangle: the proposed Antarctic Specially Protected Area). (C) Sampling areas in Inexpressible Island. Blue circle, high altitude area in Seaview Bay; red circle, low altitude area in Seaview Bay; green circle, South Bay.
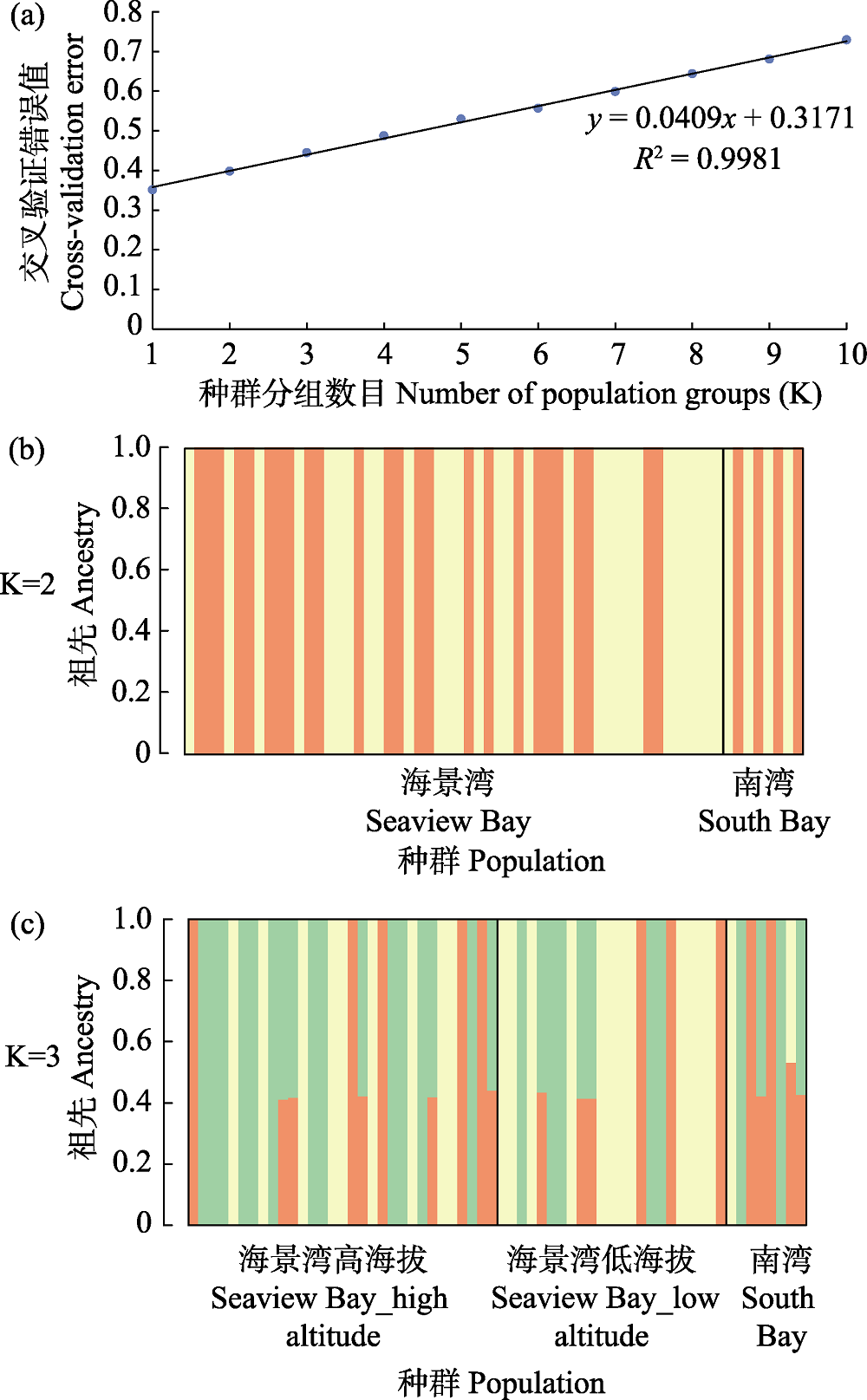
图2 基于110,451个SNPs利用最大似然法构建的恩克斯堡岛阿德利企鹅的种群遗传结构。(a)交叉验证错误率分布; (b) K = 2; (c) K = 3。图中每一竖条为1个个体, 用颜色表示每个个体基因组中不同祖先所占的比例。
Fig. 2 Based on 110,451 SNPs, the population genetic structure of Adélie penguins was constructed by maximum likelihood method. (a) Distribution of cross-validation errors; (b) K = 2 and (c) K = 3, each vertical bar represents an individual, and the colours show the proportion of ancestry assigned to each of the clusters.
| [1] | Ainley DG, LeResche RE, Sladen WJL ( 1983) Breeding Biology of the Adélie Penguin. University of California Press, Oakland. |
| [2] | Alexander DH, Novembre J, Lange K ( 2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655-1664. |
| [3] | Baroni C, Hall BL ( 2004) A new Holocene relative sea-level curve for Terra Nova Bay, Victoria Land, Antarctica. Journal of Quaternary Science, 19, 377-396. |
| [4] | Baroni C, Orombelli G ( 1991) Holocene raised beaches at Terra Nova Bay, Victoria Land, Antarctica. Quaternary Research, 36, 157-177. |
| [5] | Borowicz A, McDowall P, Youngflesh C, Sayre-McCord T, Clucas G, Herman R, Forrest S, Rider M, Schwaller M, Hart T, Jenouvrier S, Polito MJ, Singh H, Lynch HJ ( 2018) Multi-modal survey of Adélie penguin mega-colonies reveals the Danger Islands as a seabird hotspot. Scientific Reports, 8, 9. |
| [6] | Bromwich DH, Kurtz DD ( 1984) Katabatic wind forcing of the Terra Nova Bay polynya. Journal of Geophysical Research Oceans, 89, 3561-3572. |
| [7] | Clucas GV, Younger JL, Kao D, Emmerson L, Southwell C, Wienecke B, Rogers AD, Bost CA, Miller GD, Polito MJ, Lelliott P, Handley J, Crofts S, Phillips RA, Dunn MJ, Miller KJ, Hart T ( 2018) Comparative population genomics reveals key barriers to dispersal in Southern Ocean penguins. Molecular Ecology, 27, 4680-4697. |
| [8] | Clucas GV, Younger JL, Kao D, Rogers AD, Handley J, Miller GD, Jouventin P, Nolan P, Gharbi K, Miller KJ, Hart T ( 2016) Dispersal in the sub-Antarctic: King penguins show remarkably little population genetic differentiation across their range. BMC Evolutionary Biology, 16, 211. |
| [9] | Croxall JP, Prince PA ( 1979) Antarctic seabird and seal monitoring studies. Polar Record, 19, 573-595. |
| [10] | Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST ( 2011) The variant call format and VCFtools. Bioinformatics, 27, 2156-2158. |
| [11] | Emslie SD, Coats L, Licht K ( 2007) A 45,000 yr record of Adélie penguins and climate change in the Ross Sea, Antarctica. Geology, 35, 61-64. |
| [12] | Freed D, Aldana R, Weber JA, Edwards JS ( 2017) The Sentieon Genomics Tools—A fast and accurate solution to variant calling from next-generation sequence data. bioRxiv, 115717. |
| [13] | Funk WC, McKay JK, Hohenlohe PA, Allendorf FW ( 2012) Harnessing genomics for delineating conservation units. Trends in Ecology & Evolution, 27, 489-496. |
| [14] | Gorman KB, Talbot SL, Sonsthagen SA, Sage GK, Gravely MC, Fraser WR, Williams TD ( 2017) Population genetic structure and gene flow of Adélie penguins (Pygoscelis adeliae) breeding throughout the western Antarctic Peninsula. Antarctic Science, 29, 499-510. |
| [15] | Li H, Durbin R ( 2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26, 589-595. |
| [16] | Lischer HEL, Excoffier L ( 2012) PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics, 28, 298-299. |
| [17] | Lorenzini S, Baroni C, Fallick AE, Baneschi I, Salvatore MC, Zanchetta G, Dallai L ( 2010) Stable isotopes reveal Holocene changes in the diet of Adélie penguins in Northern Victoria Land (Ross Sea, Antarctica). Oecologia, 164, 911-919. |
| [18] | Lynch HJ, LaRue MA ( 2016) First global census of the Adélie penguin. Auk, 133, 236. |
| [19] | Lyver POB, Barron M, Barton KJ, Ainley DG, Pollard A, Gordon S, McNeill S, Ballard G, Wilson PR ( 2014) Trends in the breeding population of Adélie penguins in the Ross Sea, 1981-2012: A coincidence of climate and resource extraction effects. PLoS ONE, 9, e91188. |
| [20] | Manel S, Schwartz MK, Luikart G, Taberlet P ( 2003) Landscape genetics: Combining landscape ecology and population genetics. Trends in Ecology & Evolution, 18, 189-197. |
| [21] | Nielsen R, Paul JS, Albrechtsen A, Song Y ( 2011) Genotype and SNP calling from next-generation sequencing data. Nature Reviews Genetics, 12, 443-451. |
| [22] | Palsboll PJ, Berube M, Allendorf FW ( 2007) Identification of management units using population genetic data. Trends in Ecology & Evolution, 22, 11-16. |
| [23] | Peakall R, Smouse PE ( 2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics, 28, 2537-2539. |
| [24] | Raj A, Stephens M, Pritchard JK ( 2014) fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics, 197, 573-589. |
| [25] | Ritchie PA, Millar CD, Gibb GC, Baroni C, Lambert DM ( 2004) Ancient DNA enables timing of the Pleistocene origin and Holocene expansion of two Adélie penguin lineages in Antarctica. Molecular Biology and Evolution, 21, 240-248. |
| [26] | Shepherd LD, Millar CD, Ballard G, Ainley DG, Wilson PR, Haynes GD, Baroni C, Lambert DM ( 2005) Microevolution and mega-icebergs in the Antarctic. Proceedings of the National Academy of Sciences, USA, 102, 16717-16722. |
| [27] | Stonehouse B ( 1969) Air census of two colonies of Adélie penguins (Pygoscelis adeliae) in Ross Dependency, Antarctica. Polar Record, 14, 471-475. |
| [28] | Walther GR, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC, Fromentin JM, Hoegh-Guldberg O, Bairlein F ( 2002) Ecological responses to recent climate change. Nature, 416, 389-395. |
| [29] | Woehler EJ, Croxall JP ( 1997) The status and trends of Antarctic and sub-Antarctic seabirds. Marine Ornithology, 25, 43-66. |
| [30] | Younger JL, Clucas GV, Kao D, Rogers AD, Gharbi K, Hart T, Miller KJ ( 2017) The challenges of detecting subtle population structure and its importance for the conservation of emperor penguins. Molecular Ecology, 26, 3883-3897. |
| [31] | Zhang C, Dong SS, Xu JY, He WM, Yang TL ( 2019) PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics, 35, 1786-1788. |
| [32] | Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW ( 2014) Comparative genomics reveals insights into avian genome evolution and adaptation. Science, 346, 1311-1320. |
| [33] | Zheng XW, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS ( 2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics, 28, 3326-3328. |
| [1] | 邓洪, 钟占友, 寇春妮, 朱书礼, 李跃飞, 夏雨果, 武智, 李捷, 陈蔚涛. 基于线粒体全基因组揭示斑鳠的种群遗传结构与演化历史[J]. 生物多样性, 2025, 33(1): 24241-. |
| [2] | 王嘉陈, 徐汤俊, 许唯, 张高季, 尤艺瑾, 阮宏华, 刘宏毅. 城市景观格局对大蚰蜒种群遗传结构的影响[J]. 生物多样性, 2025, 33(1): 24251-. |
| [3] | 孙维悦, 舒江平, 顾钰峰, 莫日根高娃, 杜夏瑾, 刘保东, 严岳鸿. 基于保护基因组学揭示荷叶铁线蕨的濒危机制[J]. 生物多样性, 2022, 30(7): 21508-. |
| [4] | 陈向向, 盖中帅, 翟军团, 徐劲东, 焦培培, 吴智华, 李志军. 中国西北地区天然胡杨群体遗传多样性及核心保护单元的构建[J]. 生物多样性, 2021, 29(12): 1638-1649. |
| [5] | 向登高, 李跃飞, 李新辉, 陈蔚涛, 马秀慧. 多基因联合揭示海南鲌的遗传结构与遗传多样性[J]. 生物多样性, 2021, 29(11): 1505-1512. |
| [6] | 张亚红, 贾会霞, 王志彬, 孙佩, 曹德美, 胡建军. 滇杨种群遗传多样性与遗传结构[J]. 生物多样性, 2019, 27(4): 355-365. |
| [7] | 武星彤, 陈璐, 王敏求, 张原, 林雪莹, 李鑫玉, 周宏, 文亚峰. 丹霞梧桐群体遗传结构及其遗传分化[J]. 生物多样性, 2018, 26(11): 1168-1179. |
| [8] | 刘青青, 董志军. 基于线粒体COI基因分析钩手水母的群体遗传结构[J]. 生物多样性, 2018, 26(11): 1204-1211. |
| [9] | 熊敏, 田双, 张志荣, 范邓妹, 张志勇. 华木莲居群遗传结构与保护单元[J]. 生物多样性, 2014, 22(4): 476-484. |
| [10] | 杨爱红, 张金菊, 田华, 姚小洪, 黄宏文. 鹅掌楸贵州烂木山居群的微卫星遗传多样性及空间遗传结构[J]. 生物多样性, 2014, 22(3): 375-384. |
| [11] | 徐刚标, 梁艳, 蒋燚, 刘雄盛, 胡尚力, 肖玉菲, 郝博搏. 伯乐树种群遗传多样性及遗传结构[J]. 生物多样性, 2013, 21(6): 723-731. |
| [12] | 王德元, 彭婕, 陈雅静, 吕国胜, 张小平, 邵剑文. 毛茛叶报春的遗传多样性及遗传结构[J]. 生物多样性, 2013, 21(5): 601-609. |
| [13] | 李晶晶, 张杰, 胡自民, 段德麟. 加拿大-西北∇大西洋地区掌形藻的种群遗传结构与动态变化[J]. 生物多样性, 2013, 21(3): 306-314. |
| [14] | 阮咏梅, 张金菊, 姚小洪, 叶其刚. 黄梅秤锤树孤立居群的遗传多样性及其小尺度空间遗传结构[J]. 生物多样性, 2012, 20(4): 460-469. |
| [15] | 曹玲亮, 周立志, 张保卫. 安徽三大水系入侵物种克氏原螯虾的种群遗传格局[J]. 生物多样性, 2010, 18(4): 398-407. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn
![]()