



Biodiv Sci ›› 2024, Vol. 32 ›› Issue (9): 24212. DOI: 10.17520/biods.2024212 cstr: 32101.14.biods.2024212
• Genetic Diversity • Previous Articles Next Articles
Xiangtan Yao1, Xinyi Zhang2,3, Yang Chen4, Ye Yuan1, Wangda Cheng5,*( ), Tianrui Wang2,6,*(
), Tianrui Wang2,6,*( )(
)( ), Yingxiong Qiu2,*(
), Yingxiong Qiu2,*( )(
)( )
)
Received:2024-05-30
Accepted:2024-08-01
Online:2024-09-20
Published:2024-09-02
Contact:
* E-mail: chwd228@163.com;
wangtianrui231@mails.ucas.ac.cn;
qiuyingxiong@wbgcas.cnSupported by:Xiangtan Yao, Xinyi Zhang, Yang Chen, Ye Yuan, Wangda Cheng, Tianrui Wang, Yingxiong Qiu. Genomic resequencing reveals the genetic diversity of the cultivated water caltrop, and the origin and domestication of ‘Nanhuling’[J]. Biodiv Sci, 2024, 32(9): 24212.
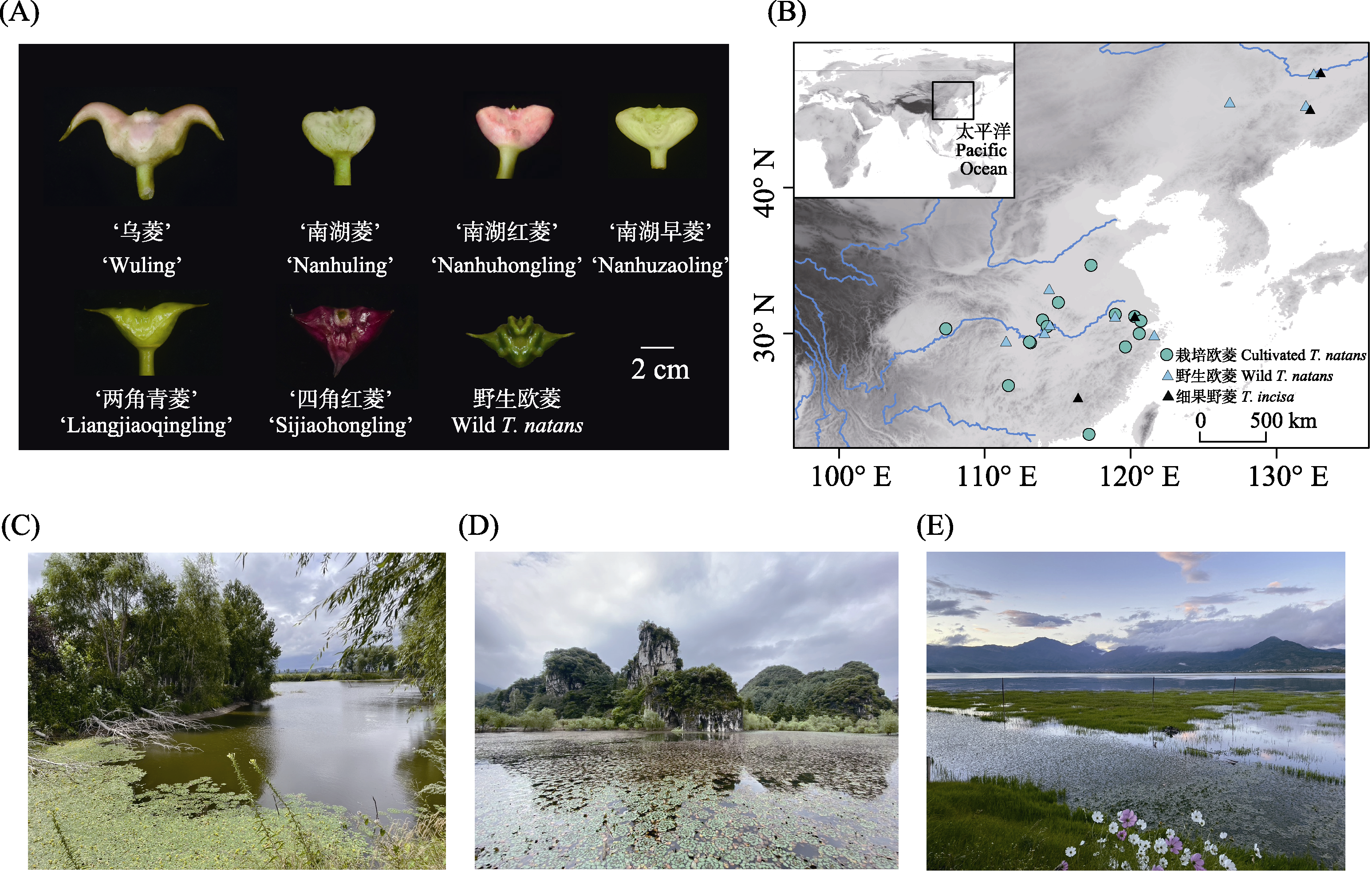
Fig. 1 Fruit morphology, collection site, and distribution habitat of cultivated and wild Trapa natans. A, Fruit morphology; B, Collection site; C-E, Typical habitat of Trapa.
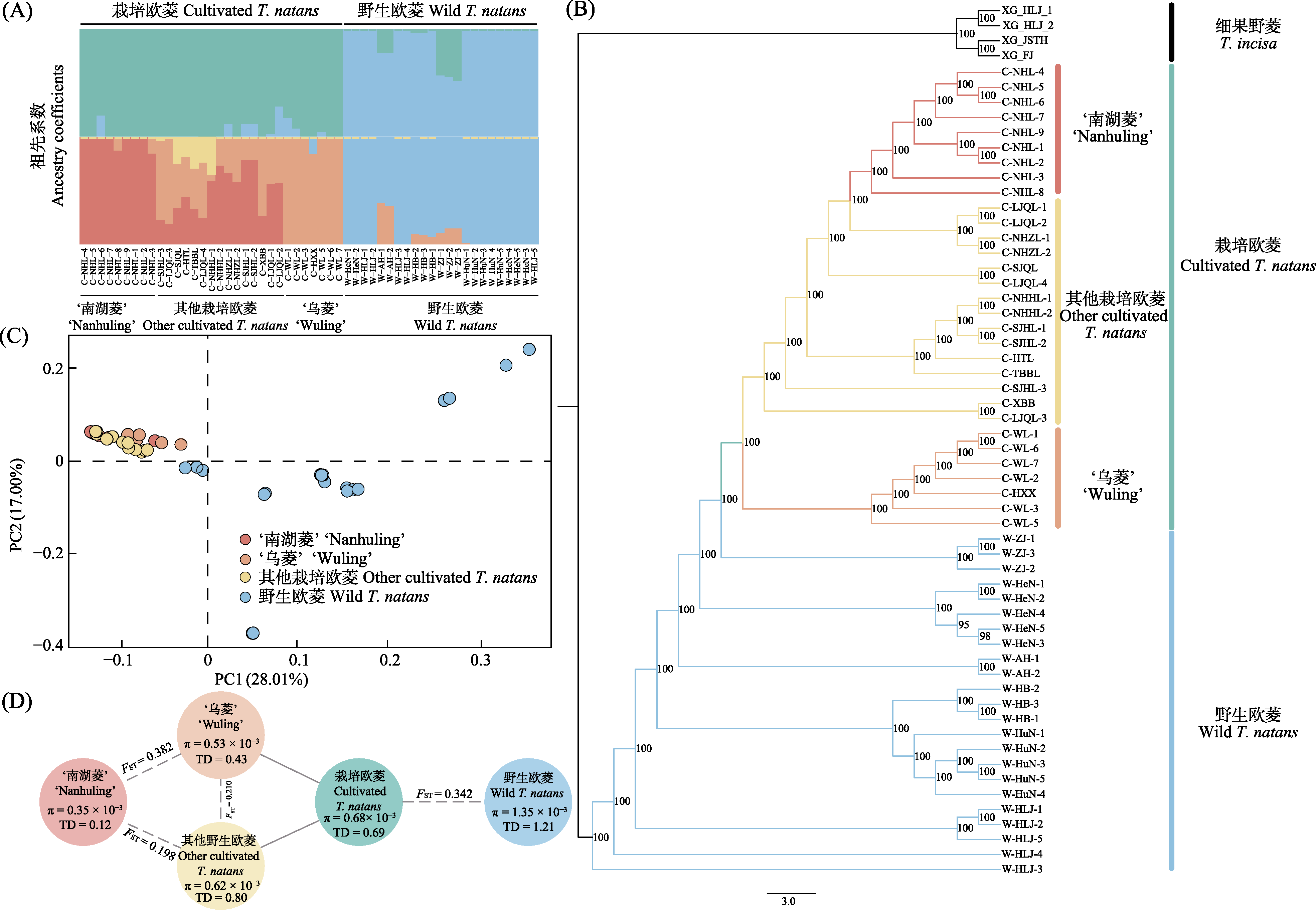
Fig. 2 Population genetic structure and divergence of cultivated and wild Trapa natans. A, Population genetic structure based on ADMIXTURE analysis; B, Maximum likelihood (ML) phylogenetic tree; C, Principal component analysis (PCA); D, Estimates of nucleotide diversity (π) and Tajima’s D (TD) and genetic differentiation index (FST) for wild and cultivated T. natans.
| 模型 Models | 参数Parameters | Delta似然值 Delta likelihood | AIC | AIC权重 AIC’s weight |
|---|---|---|---|---|
| Model 1 | 23 | 55,516.96 | 2,597,573.64 | 1 |
| Model 2 | 21 | 56,021.93 | 2,640,483.74 | 0 |
| Model 3 | 19 | 59,304.03 | 2,615,594.33 | 0 |
| Model 4 | 17 | 71,677.36 | 2,672,571.65 | 0 |
| Model 5 | 15 | 69,289.32 | 2,661,570.30 | 0 |
| Model 6 | 21 | 55,952.22 | 2,610,162.71 | 0 |
| Model 7 | 17 | 67,543.76 | 2,653,535.73 | 0 |
| Model 8 | 19 | 65,390.88 | 2,698,162.24 | 0 |
Table 1 Likelihood comparison of models for cultivated and wild Trapa natans based on simulations using FASTSIMCOAL2
| 模型 Models | 参数Parameters | Delta似然值 Delta likelihood | AIC | AIC权重 AIC’s weight |
|---|---|---|---|---|
| Model 1 | 23 | 55,516.96 | 2,597,573.64 | 1 |
| Model 2 | 21 | 56,021.93 | 2,640,483.74 | 0 |
| Model 3 | 19 | 59,304.03 | 2,615,594.33 | 0 |
| Model 4 | 17 | 71,677.36 | 2,672,571.65 | 0 |
| Model 5 | 15 | 69,289.32 | 2,661,570.30 | 0 |
| Model 6 | 21 | 55,952.22 | 2,610,162.71 | 0 |
| Model 7 | 17 | 67,543.76 | 2,653,535.73 | 0 |
| Model 8 | 19 | 65,390.88 | 2,698,162.24 | 0 |
| 参数 Parameters | 点估计 Point estimation | 95%置信区间 95% confidence intervals | |
|---|---|---|---|
| 置信下限 Lower bound | 置信上限 Upper bound | ||
| 野生欧菱有效群体大小 Effective population size of wild T. natans | 7,685 | 3,873 | 10,423 |
| ‘乌菱’有效群体大小 Effective population size of ‘Wuling’ | 1,465 | 770 | 2,128 |
| ‘南湖菱’有效群体大小 Effective population size of ‘Nanhuling’ | 1,087 | 586 | 2,526 |
| 其他栽培欧菱有效群体大小 Effective population size of other cultivated T. natans | 4,823 | 3,088 | 6,492 |
| 栽培欧菱祖先有效群体大小 Effective population size of the ancestor of cultivated T. natans | 4,068 | 3,293 | 7,602 |
| ‘南湖菱’和其他栽培欧菱的祖先有效群体大小 Effective population size of the ancestor of ‘Nanhuling’ and other cultivated T. natans | 5,245 | 4,995 | 8,423 |
| 欧菱祖先有效群体大小 Effective population size of the ancestor of T. natans | 31,528 | 18,113 | 42,355 |
| ‘南湖菱’驯化时间 Domestication time of ‘Nanhuling’ | 669 | 285 | 983 |
| ‘乌菱’驯化时间 Domestication time of ‘Wuling’ | 839 | 661 | 1,428 |
| 栽培欧菱驯化时间 Domestication time of cultivated T. natans | 6,741 | 4,658 | 8,237 |
| ‘南湖菱’向‘乌菱’迁移率 Migration rate from ‘Nanhuling’ to ‘Wuling’ | 6.68E-6 | 3.27E-6 | 4.12E-5 |
| ‘乌菱’向‘南湖菱’迁移率 Migration rate from ‘Wuling’ to ‘Nanhuling’ | 1.16E-5 | 6.67E-6 | 4.82E-5 |
| 其他栽培欧菱向‘南湖菱’迁移率 Migration rate from other cultivated T. natans to ‘Nanhuling’ | 2.29E-5 | 3.31E-6 | 4.44E-5 |
| ‘南湖菱’向其他栽培欧菱迁移率 Migration rate from ‘Nanhuling’ to other cultivated T. natans | 3.40E-5 | 4.67E-6 | 9.73E-5 |
| ‘南湖菱’和其他栽培欧菱祖先向‘乌菱’迁移率 Migration rate from the ancestor of ‘Nanhuling’ and other cultivated T. natans to ‘Wuling’ | 1.52E-3 | 7.24E-4 | 4.29E-3 |
| ‘乌菱’向‘南湖菱’和其他栽培欧菱祖先迁移率 Migration rate from ‘Wuling’ to the ancestor of ‘Nanhuling’ and other cultivated T. natans | 1.27E-3 | 6.28E-4 | 4.64E-3 |
| 野生欧菱向栽培欧菱迁移率 Migration rate from wild T. natans to cultivated T. natans | 9.25E-3 | 2.92E-3 | 1.76E-2 |
| 栽培欧菱向野生欧菱迁移率 Migration rate from cultivated T. natans to wild T. natans | 1.75E-4 | 7.66E-5 | 1.13E-3 |
| 其他栽培欧菱向‘乌菱’迁移率 Migration rate from other cultivated T. natans to ‘Wuling’ | 1.95E-6 | 2.58E-7 | 8.69E-5 |
| ‘乌菱’向其他栽培欧菱迁移率 Migration rate from ‘Wuling’ to other cultivated T. natans | 3.70E-6 | 3.31E-6 | 1.35E-5 |
Table 2 Parameter estimates of effective population sizes, domestication time, and migration events for cultivated and wild Trapa natans
| 参数 Parameters | 点估计 Point estimation | 95%置信区间 95% confidence intervals | |
|---|---|---|---|
| 置信下限 Lower bound | 置信上限 Upper bound | ||
| 野生欧菱有效群体大小 Effective population size of wild T. natans | 7,685 | 3,873 | 10,423 |
| ‘乌菱’有效群体大小 Effective population size of ‘Wuling’ | 1,465 | 770 | 2,128 |
| ‘南湖菱’有效群体大小 Effective population size of ‘Nanhuling’ | 1,087 | 586 | 2,526 |
| 其他栽培欧菱有效群体大小 Effective population size of other cultivated T. natans | 4,823 | 3,088 | 6,492 |
| 栽培欧菱祖先有效群体大小 Effective population size of the ancestor of cultivated T. natans | 4,068 | 3,293 | 7,602 |
| ‘南湖菱’和其他栽培欧菱的祖先有效群体大小 Effective population size of the ancestor of ‘Nanhuling’ and other cultivated T. natans | 5,245 | 4,995 | 8,423 |
| 欧菱祖先有效群体大小 Effective population size of the ancestor of T. natans | 31,528 | 18,113 | 42,355 |
| ‘南湖菱’驯化时间 Domestication time of ‘Nanhuling’ | 669 | 285 | 983 |
| ‘乌菱’驯化时间 Domestication time of ‘Wuling’ | 839 | 661 | 1,428 |
| 栽培欧菱驯化时间 Domestication time of cultivated T. natans | 6,741 | 4,658 | 8,237 |
| ‘南湖菱’向‘乌菱’迁移率 Migration rate from ‘Nanhuling’ to ‘Wuling’ | 6.68E-6 | 3.27E-6 | 4.12E-5 |
| ‘乌菱’向‘南湖菱’迁移率 Migration rate from ‘Wuling’ to ‘Nanhuling’ | 1.16E-5 | 6.67E-6 | 4.82E-5 |
| 其他栽培欧菱向‘南湖菱’迁移率 Migration rate from other cultivated T. natans to ‘Nanhuling’ | 2.29E-5 | 3.31E-6 | 4.44E-5 |
| ‘南湖菱’向其他栽培欧菱迁移率 Migration rate from ‘Nanhuling’ to other cultivated T. natans | 3.40E-5 | 4.67E-6 | 9.73E-5 |
| ‘南湖菱’和其他栽培欧菱祖先向‘乌菱’迁移率 Migration rate from the ancestor of ‘Nanhuling’ and other cultivated T. natans to ‘Wuling’ | 1.52E-3 | 7.24E-4 | 4.29E-3 |
| ‘乌菱’向‘南湖菱’和其他栽培欧菱祖先迁移率 Migration rate from ‘Wuling’ to the ancestor of ‘Nanhuling’ and other cultivated T. natans | 1.27E-3 | 6.28E-4 | 4.64E-3 |
| 野生欧菱向栽培欧菱迁移率 Migration rate from wild T. natans to cultivated T. natans | 9.25E-3 | 2.92E-3 | 1.76E-2 |
| 栽培欧菱向野生欧菱迁移率 Migration rate from cultivated T. natans to wild T. natans | 1.75E-4 | 7.66E-5 | 1.13E-3 |
| 其他栽培欧菱向‘乌菱’迁移率 Migration rate from other cultivated T. natans to ‘Wuling’ | 1.95E-6 | 2.58E-7 | 8.69E-5 |
| ‘乌菱’向其他栽培欧菱迁移率 Migration rate from ‘Wuling’ to other cultivated T. natans | 3.70E-6 | 3.31E-6 | 1.35E-5 |
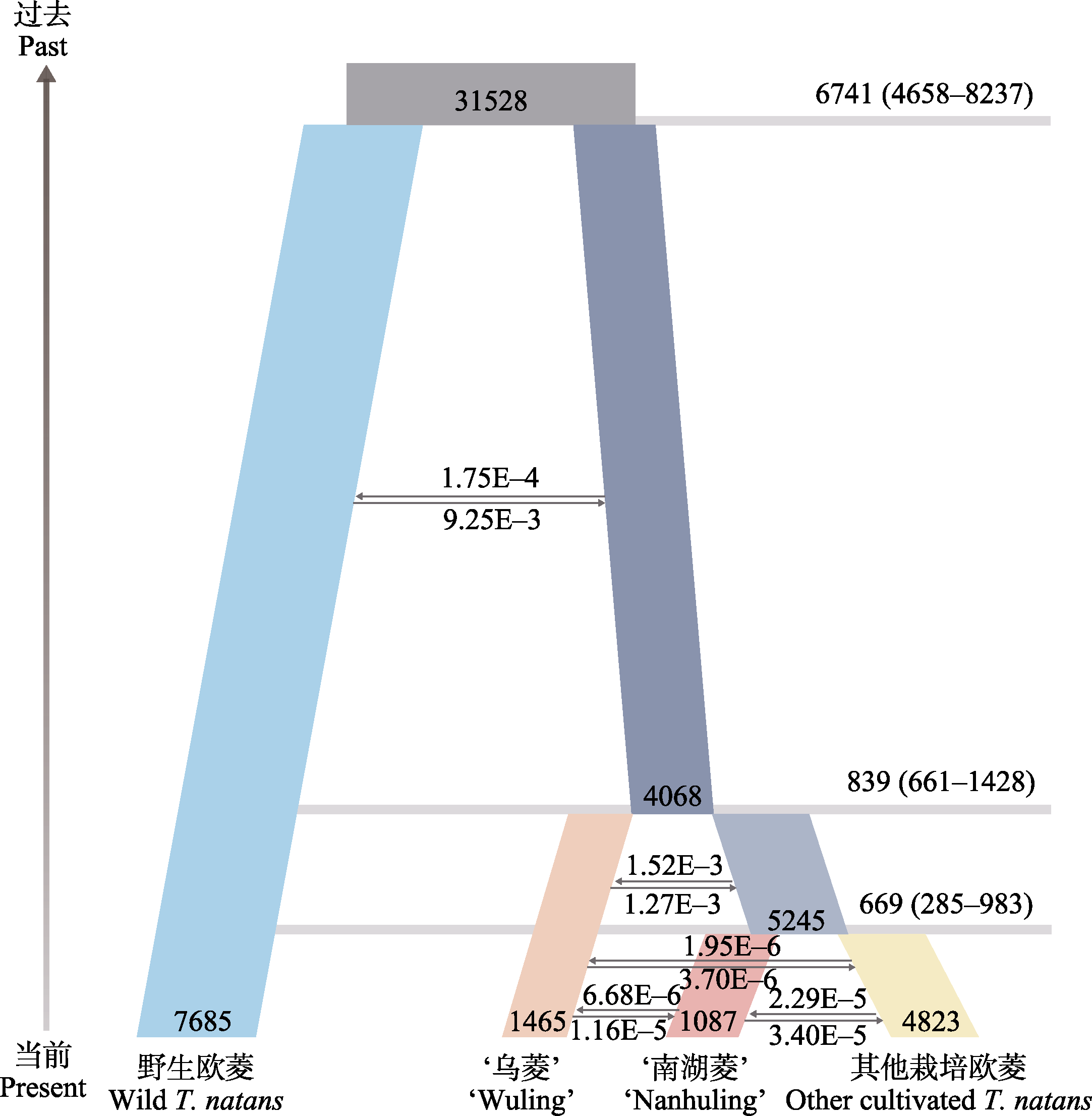
Fig. 4 Schematic of the best demographic scenario modeled using FASTSIMCOAL2. The numbers on the right axis indicate the domestication time (year) before present and 95% confidence interval. Column width represents the relative effective population size. The arrows and the numbers above them represent gene flow. The bars in different colors represent different Trapa natans.
| [1] | Agapow PM, Burt A (2001) Indices of multilocus linkage disequilibrium. Molecular Ecology Notes, 1, 101-102. |
| [2] |
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Research, 19, 1655-1664.
DOI PMID |
| [3] | Bosse M, Megens HJ, Derks MFL, de Cara ÁMR, Groenen MAM (2018) Deleterious alleles in the context of domestication, inbreeding, and selection. Evolutionary Applications, 12, 6-17. |
| [4] | Bouzat JL (2010) Conservation genetics of population bottlenecks: The role of chance, selection, and history. Conservation Genetics, 11, 463-478. |
| [5] |
Chapman MA, He YQ, Zhou ML (2022) Beyond a reference genome: Pangenomes and population genomics of underutilized and orphan crops for future food and nutrition security. New Phytologist, 234, 1583-1597.
DOI PMID |
| [6] | Chang Y, Liu H, Liu M, Liao XZ, Sahu SK, Fu Y, Song B, Cheng SF, Kariba R, Muthemba S, Hendre PS, Mayes S, Ho WK, Yssel AEJ, Kendabie P, Wang SB, Li LZ, Muchugi A, Jamnadass R, Lu HR, Peng SF, Van Deynze A, Simons A, Yana-Shapiro H, Van de Peer Y, Xu X, Yang HM, Wang J, Liu X (2019) The draft genomes of five agriculturally important African orphan crops. GigaScience, 8, giy152. |
| [7] | Chen Y (2023) Allopolyploid Formation in Trapa, and the Origin and Domestication of Water Caltrop. PhD dissertation, Zhejiang University, Hangzhou. (in Chinese with English abstract) |
| [陈阳 (2023) 菱属植物异源多倍体形成及栽培欧菱的起源驯化研究. 博士学位论文, 浙江大学, 杭州.] | |
| [8] |
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, Genomes Project Analysis Group 1000 (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156-2158.
DOI PMID |
| [9] | Ding BY, Hu RY, Shi MZ, Zheng CZ (1996) A preliminary study on the pollination biology of Trapa L. Journal of Hangzhou University (Natural Science), 23, 275-279. (in Chinese with English abstract) |
| [丁炳扬, 胡仁勇, 史美中, 郑朝宗 (1996) 菱属植物传粉生物学的初步研究. 杭州大学学报(自然科学版), 23, 275-279.] | |
| [10] | Ding BY, Jin XF (2020) Taxonomic notes on genus Trapa L. (Trapaceae) in China. Guihaia, 40, 1-15. |
| [11] |
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell, 127, 1309-1321.
DOI PMID |
| [12] | Doebley J (1989) Isozymic evidence and the evolution of crop plants. In: Isozymes in Plant Biology (eds Soltis DE, Soltis PS, Dudley TR), pp.165-191. Springer, Dordrecht. |
| [13] |
Ellegren H, Galtier N (2016) Determinants of genetic diversity. Nature Reviews Genetics, 17, 422-433.
DOI PMID |
| [14] | Excoffier L, Dupanloup I, Huerta-Sánchez E, Sousa VC, Foll M (2013) Robust demographic inference from genomic and SNP data. PLoS Genetics, 9, e1003905. |
| [15] |
Excoffier L, Foll M (2011) Fastsimcoal: A continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics, 27, 1332-1334.
DOI PMID |
| [16] |
Fuller DQ (2007) Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the old world. Annals of Botany, 100, 903-924.
DOI PMID |
| [17] |
Gaut BS, Díez CM, Morrell PL (2015) Genomics and the contrasting dynamics of annual and perennial domestication. Trends in Genetics, 31, 709-719.
DOI PMID |
| [18] |
Gaut BS, Seymour DK, Liu QP, Zhou YF (2018) Demography and its effects on genomic variation in crop domestication. Nature Plants, 4, 512-520.
DOI PMID |
| [19] | Gepts P (2003) Crop domestication as a long-term selection experiment. Plant Breeding Reviews, 24, 1-44. |
| [20] | Gross BL, Zhao ZJ (2014) Archaeological and genetic insights into the origins of domesticated rice. Proceedings of the National Academy of Sciences, USA, 111, 6190-6197. |
| [21] |
Guerra-García A, Rojas-Barrera IC, Ross-Ibarra J, Papa R, Piñero D (2022) The genomic signature of wild-to-crop introgression during the domestication of scarlet runner bean (Phaseolus coccineus L.). Evolution Letters, 6, 295-307.
DOI PMID |
| [22] | Guo C, Wang YN, Yang AG, He J, Xiao CW, Lv SH, Han FM, Yuan YB, Yuan Y, Dong XL, Guo J, Yang YW, Liu HL, Zuo NZ, Hu YX, Zhao KX, Jiang ZB, Wang X, Jiang TT, Shen YO, Cao MJ, Wang Y, Long ZB, Rong TZ, Huang LQ, Zhou SF (2020) The Coix genome provides insights into Panicoideae evolution and papery hull domestication. Molecular Plant, 13, 309-320. |
| [23] |
Guo Y, Wu RB, Sun GP, Zheng YF, Fuller BT (2017) Neolithic cultivation of water chestnuts (Trapa L.) at Tianluoshan (7000-6300 cal BP), Zhejiang Province, China. Scientific Reports, 7, 16206.
DOI PMID |
| [24] |
He YQ, Zhang KX, Shi YL, Lin H, Huang X, Lu X, Wang ZR, Li W, Feng XB, Shi TX, Chen QF, Wang JZ, Tang Y, Chapman MA, Germ M, Luthar Z, Kreft I, Janovská D, Meglič V, Woo SH, Quinet M, Fernie AR, Liu X, Zhou ML (2024) Genomic insight into the origin, domestication, dispersal, diversification and human selection of Tartary buckwheat. Genome Biology, 25, 61.
DOI PMID |
| [25] | Hoque A, Davey MR, Arima S (2009) Water chestnut: Potential of biotechnology for crop improvement. Journal of New Seeds, 10, 180-195. |
| [26] |
Huang R, Liu YR, Chen JL, Lu ZY, Wang JJ, He W, Chao Z, Tian EW (2022) Limited genetic diversity and high differentiation in Angelica dahurica resulted from domestication: Insights to breeding and conservation. BMC Plant Biology, 22, 141.
DOI PMID |
| [27] | Huang XH, Kurata N, Wei XH, Wang ZX, Wang AH, Zhao Q, Zhao Y, Liu KY, Lu HY, Li WJ, Guo YL, Lu YQ, Zhou CC, Fan DL, Weng QJ, Zhu CR, Huang T, Zhang L, Wang YC, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan XP, Xu Q, Dong GJ, Zhan QL, Li CY, Fujiyama A, Toyoda A, Lu TT, Feng Q, Qian Q, Li JY, Han B (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature, 490, 497-501. |
| [28] | Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia JM, Cartwright RA, Elshire RJ, Glaubitz JC, Guill KE, Kaeppler SM, Lai JS, Morrell PL, Shannon LM, Song C, Springer NM, Swanson-Wagner RA, Tiffin P, Wang J, Zhang GY, Doebley J, McMullen MD, Ware D, Buckler ES, Yang S, Ross-Ibarra J (2012) Comparative population genomics of maize domestication and improvement. Nature Genetics, 44, 808-811. |
| [29] | Hummel M, Kiviat E (2004) Review of world literature on water chestnut with implications for management in North America. Journal of Aquatic Plant Management, 42, 17-28. |
| [30] | Jarvis DE, Ho YS, Lightfoot DJ, Schmöckel SM, Li B, Borm TJA, Ohyanagi H, Mineta K, Michell CT, Saber N, Kharbatia NM, Rupper RR, Sharp AR, Dally N, Boughton BA, Woo YH, Gao G, Schijlen EGWM, Guo XJ, Momin AA, Negrão S, Al-Babili S, Gehring C, Roessner U, Jung C, Murphy K, Arold ST, Gojobori T, van der Linden CG, van Loo EN, Jellen EN, Maughan PJ, Tester M (2017) The genome of Chenopodium quinoa. Nature, 542, 307-312. |
| [31] |
Jombart T, Ahmed I (2011) Adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics, 27, 3070-3071.
DOI PMID |
| [32] | Karg S (2006) The water chestnut (Trapa natans L.) as a food resource during the 4th to 1st millennia BC at Lake Federsee, Bad Buchau (southern Germany). Environmental Archaeology, 11, 125-130. |
| [33] | Kim C, Na HR, Choi HK (2010) Molecular genotyping of Trapa bispinosa and T. japonica (Trapaceae) based on nuclear AP2 and chloroplast DNA trnL-F region. American Journal of Botany, 97, e149-e152. |
| [34] |
Korunes KL, Samuk K (2021) Pixy: Unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Molecular Ecology Resources, 21, 1359-1368.
DOI PMID |
| [35] | Lin QS, Chen Y, Yu HN, Shen YZ, Zeng SM, Shen SR (2013) Bioactivities of different polar fractions from three water caltrop pericarps. Science and Technology of Food Industry, 34, 139-142. (in Chinese with English abstract) |
| [林秋生, 陈莹, 于海宁, 沈钰珍, 曾思敏, 沈生荣 (2013) 三种菱壳不同极性部位提取物的生物活性. 食品工业科技, 34, 139-142.] | |
| [36] |
Lin T, Zhu GT, Zhang JH, Xu XY, Yu QH, Zheng Z, Zhang ZH, Lun YY, Li S, Wang XX, Huang ZJ, Li JM, Zhang CZ, Wang TT, Zhang YY, Wang AX, Zhang YC, Lin K, Li CY, Xiong GS, Xue YB, Mazzucato A, Causse M, Fei ZJ, Giovannoni JJ, Chetelat RT, Zamir D, Städler T, Li JF, Ye ZB, Du YC, Huang SW (2014) Genomic analyses provide insights into the history of tomato breeding. Nature Genetics, 46, 1220-1226.
DOI PMID |
| [37] |
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754-1760.
DOI PMID |
| [38] | Li XL, Fan XR, Chu HJ, Li W, Chen YY (2017) Genetic delimitation and population structure of three Trapa taxa from the Yangtze River, China. Aquatic Botany, 136, 61-70. |
| [39] |
Lowe A, Boshier D, Ward M, Bacles CFE, Navarro C (2005) Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity, 95, 255-273.
PMID |
| [40] | Lu Q, Zhao HN, Zhang ZQ, Bai YH, Zhao HM, Liu GQ, Liu MX, Zheng YX, Zhao HY, Gong HH, Chen LW, Deng XZ, Hong XD, Liu TX, Li BC, Lu P, Wen F, Wang L, Li ZJ, Li H, Li HQ, Zhang L, Ma WH, Liu CQ, Bai Y, Xin BB, Chen J, Lizhu E, Lai JS, Song WB (2024) Genomic variation in weedy and cultivated broomcorn millet accessions uncovers the genetic architecture of agronomic traits. Nature Genetics, 56, 1006-1017. |
| [41] | Lu RS, Chen Y, Zhang XY, Feng Y, Comes HP, Li Z, Zheng ZS, Yuan Y, Wang LY, Huang ZJ, Guo Y, Sun GP, Olsen KM, Chen J, Qiu YX (2022) Genome sequencing and transcriptome analyses provide insights into the origin and domestication of water caltrop (Trapa spp., Lythraceae). Plant Biotechnology Journal, 20, 761-776. |
| [42] | Meyer RS, Choi JY, Sanches M, Plessis A, Flowers JM, Amas J, Dorph K, Barretto A, Gross B, Fuller DQ, Bimpong IK, Ndjiondjop MN, Hazzouri KM, Gregorio GB, Purugganan MD (2016) Domestication history and geographical adaptation inferred from a SNP map of African rice. Nature Genetics, 48, 1083-1088. |
| [43] |
Meyer RS, Purugganan MD (2013) Evolution of crop species: Genetics of domestication and diversification. Nature Reviews Genetics, 14, 840-852.
DOI PMID |
| [44] |
Meyer RS, DuVal AE, Jensen HR (2012) Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytologist, 196, 29-48.
DOI PMID |
| [45] | Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268-274. |
| [46] |
Nielsen R (2000) Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics, 154, 931-942.
DOI PMID |
| [47] | Ning Y (2012) The Research on Bioactive Compounds against Gastric Cancer from Water Chestnut Shell. PhD dissertation, Zhejiang University, Hangzhou. (in Chinese with English abstract) |
| [宁颖 (2012) 南湖菱菱壳中抗胃癌活性成分的研究. 博士学位论文, 浙江大学, 杭州.] | |
| [48] |
Olsen KM, Wendel JF (2013) A bountiful harvest: Genomic insights into crop domestication phenotypes. Annual Review of Plant Biology, 64, 47-70.
DOI PMID |
| [49] | Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics, 81, 559-575. |
| [50] |
Purugganan MD (2019) Evolutionary insights into the nature of plant domestication. Current Biology, 29, 705-714.
DOI PMID |
| [51] |
Qi JJ, Liu X, Shen D, Miao H, Xie BY, Li XX, Zeng P, Wang SH, Shang Y, Gu XF, Du YC, Li Y, Lin T, Yuan JH, Yang XY, Chen JF, Chen HM, Xiong XY, Huang K, Fei ZJ, Mao LY, Tian L, Städler T, Renner SS, Kamoun S, Lucas WJ, Zhang ZH, Huang SW (2013) A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nature Genetics, 45, 1510-1515.
DOI PMID |
| [52] | Skaria R, Sen S, Muneer PMA (2011). Analysis of genetic variability in rice varieties (Oryza sativa L) of Kerala using RAPD markers. Genetic Engineering and Biotechnology Journal, 24, 823-831. |
| [53] | Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. (2013) From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Current Protocols in Bioinformatics, 43, 1-33. |
| [54] | von Wettberg EJB, Chang PL, Başdemir F, Carrasquila-Garcia N, Korbu LB, Moenga SM, Bedada G, Greenlon A, Moriuchi KS, Singh V, Cordeiro MA, Noujdina NV, Dinegde KN, Sani SGAS, Getahun T, Vance L, Bergmann E, Lindsay D, Mamo BE, Warschefsky EJ, Dacosta-Calheiros E, Marques E, Yilmaz MA, Cakmak A, Rose J, Migneault A, Krieg CP, Saylak S, Temel H, Friesen ML, Siler E, Akhmetov Z, Ozcelik H, Kholova J, Can CN, Gaur P, Yildirim M, Sharma H, Vadez V, Tesfaye K, Woldemedhin AF, Tar’an B, Aydogan A, Bukun B, Penmetsa RV, Berger J, Kahraman A, Nuzhdin SV, Cook DR (2018) Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nature Communications, 9, 649. |
| [55] | Wei XZ, Jiang MX (2012) Limited genetic impacts of habitat fragmentation in an “old rare” relict tree, Euptelea pleiospermum (Eupteleaceae). Plant Ecology, 213, 909-917. |
| [56] | Wu ZY, Lu AM, Tang YC, Chen ZD, Li DZ (2003) The families and genera of angiosperms in China. Science Press, Beijing. (in Chinese) |
| [吴征镒, 路安民, 汤彦承, 陈之端, 李德铢 (2003) 中国被子植物科属综论. 科学出版社, 北京.] | |
| [57] | Yao XT, Zhang M, Shen YQ (2016) Research progress of ‘Nanhuling’. Journal of Zhejiang Agricultural Sciences, 57, 1735-1739. (in Chinese) |
| [姚祥坦, 张敏, 沈亚强 (2016) 南湖菱研究进展. 浙江农业科学, 57, 1735-1739.] | |
| [58] | Ye CY, Fan LJ (2021) Orphan crops and their wild relatives in the genomic era. Molecular Plant, 14, 27-39. |
| [59] | Young AG, Merriam HG, Warwick SI (1993) The effects of forest fragmentation on genetic variation in Acer saccharum Marsh. (sugar maple) populations. Heredity, 71, 277-289. |
| [60] | Zhang XY, Chen Y, Wang LY, Yuan Y, Fang MY, Shi L, Lu RS, Comes HP, Ma YZ, Chen YY, Huang GZ, Zhou YF, Zheng ZS, Qiu YX (2023) Pangenome of water caltrop reveals structural variations and asymmetric subgenome divergence after allopolyploidization. Horticulture Research, 10, uhad203. |
| [61] |
Zhao Y, Chen JK (2018) The origin of crops in the Yangtze River Basin and its relevance for biodiversity. Biodiversity Science, 26, 333-345. (in Chinese with English abstract)
DOI |
|
[赵耀, 陈家宽 (2018) 长江流域农作物起源及其与生物多样性特征的关联. 生物多样性, 26, 333-345.]
DOI |
|
| [62] | Zhou Q (2012) The planting of water chestnut and wetland agriculture development in the Huzhou Plain in the Tang-Song dynasties. Agricultural History of China, 31(3), 11-21. (in Chinese with English abstract) |
| [周晴 (2012) 唐宋时期湖州平原菱的种植与湿地农业开发. 中国农史, 31(3), 11-21.] |
| [1] | Zhaoyang Jing, Keguang Cheng, Heng Shu, Yongpeng Ma, Pingli Liu. Whole genome resequencing approach for conservation biology of endangered plants [J]. Biodiv Sci, 2023, 31(5): 22679-. |
| [2] | Rui Luo, Ya Chen, Hanma Zhang. Research progress on whole-genome resequencing in Brassica [J]. Biodiv Sci, 2023, 31(10): 23237-. |
| [3] | Wang Xiaofan, ChenJiakuan. A survey on tbe habitats of rare and endangered aquatic plants in Hunan Province [J]. Biodiv Sci, 1994, 02(4): 193-198. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()