



Biodiv Sci ›› 2014, Vol. 22 ›› Issue (1): 51-65. DOI: 10.3724/SP.J.1003.2014.13169 cstr: 32101.14.SP.J.1003.2014.13169
Special Issue: 基因组和生物多样性
• Orginal Article • Previous Articles Next Articles
Received:2013-07-19
Accepted:2013-10-14
Online:2014-01-20
Published:2014-02-10
Contact:
Chen Mingsheng
Tieyan Liu,Mingsheng Chen. Genome evolution of Oryza[J]. Biodiv Sci, 2014, 22(1): 51-65.
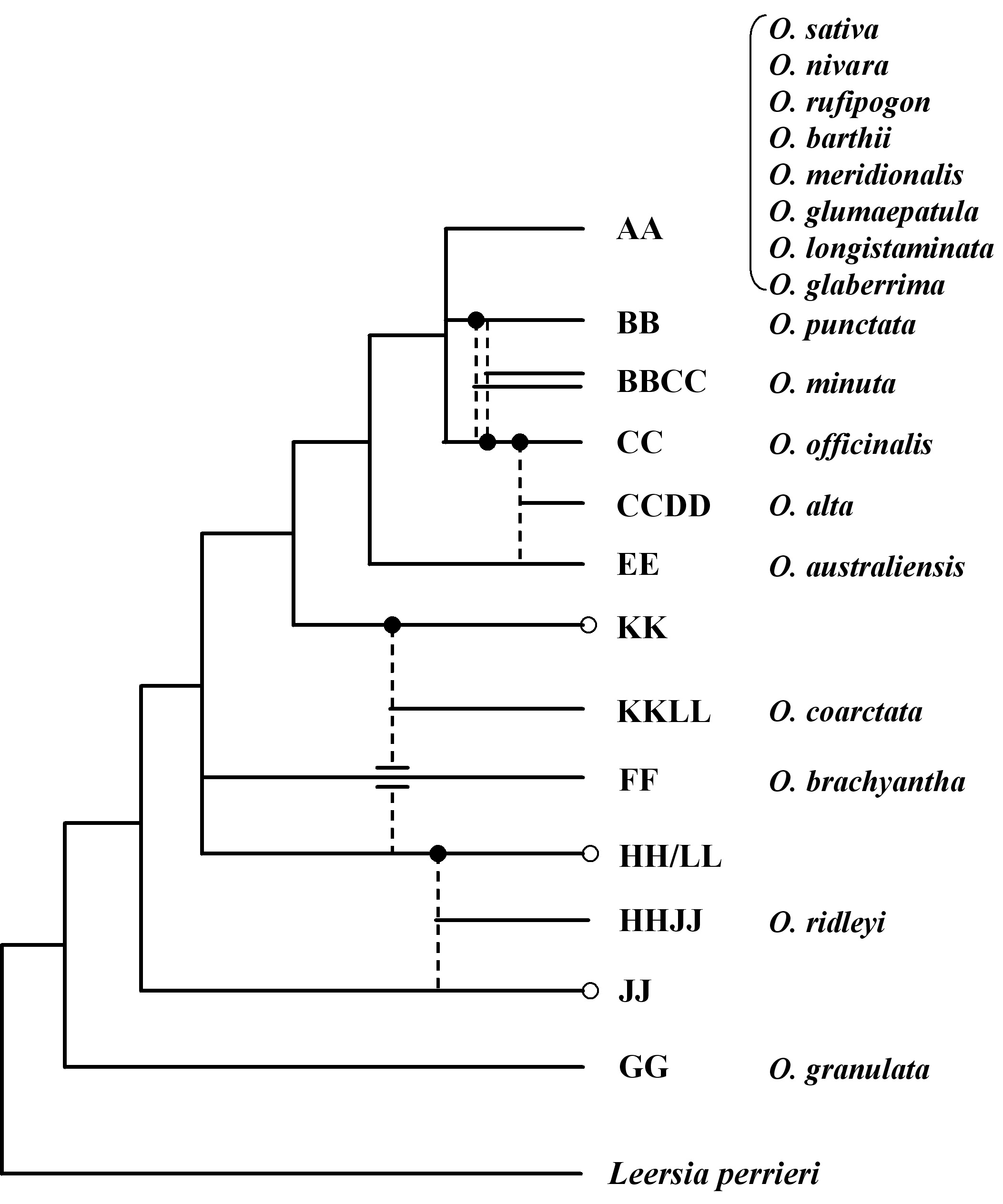
Fig. 1 Phylogenetic tree of Oryza. Dashed lines indicate putative diploid parents involved in the formation of polyploids. Filled in circles indicate maternal parent, and unfilled circles indicate unidentified diploid genome. Leersia perrieri represents the outgroup. Seventeen representative Oryza species that are currently subjected to genomic investigation as part of the International Oryza Map Alignment Project (OMAP) are indicated next to respective phylogenetic branches.
| [1] | Ahn SN, Kwon SJ, Suh JP, Kang KH, Kim HJ, Hwang HG (2002) Introgressions of Oryza grandiglumis chromatin into rice affect plant height and grain length.Rice Genetics Newsletter, 19, 12-13. |
| [2] | Ali ML, Sanchez PL, Yu SB, Lorieux M, Eizenga GC (2010) Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa).Rice, 3, 218-234. |
| [3] | Amante-Bordeos A, Sitch LA, Nelson R, Dalmacio RD, Oliva NP, Aswidinnoor H, Leung H (1992) Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa.Theoretical and Applied Genetics, 84, 345-354. |
| [4] | Ammiraju JS, Fan CZ, Yu Y, Song X, Cranston KA, Pontaroli AC, Lu F, Sanyal A, Jiang N, Rambo T, Currie J, Collura K, Talag J, Bennetzen JL, Chen MS, Jackson S, Wing RA (2010) Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza.The Plant Journal, 63, 430-442. |
| [5] | Ammiraju JS, Lu F, Sanyal A, Yu Y, Song X, Jiang N, Pontaroli AC, Rambo T, Currie J, Collura K, Talag J, Fan C, Goicoechea JL, Zuccolo A, Chen J, Bennetzen JL, Chen M, Jackson S, Wing RA (2008) Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set.The Plant Cell, 20, 3191-3209. |
| [6] | Ammiraju JS, Luo M, Goicoechea JL, Wang W, Kudrna D, Mueller C, Talag J, Kim H, Sisneros NB, Blackmon B, Fang E, Tomkins JB, Brar D, MacKill D, McCouch S, Kurata N, Lambert G, Galbraith DW, Arumuganathan K, Rao K, Walling JG, Gill N, Yu Y, SanMiguel P, Soderlund C, Jackson S, Wing RA (2006) The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza.Genome Research, 16, 140-147. |
| [7] | Ammiraju JS, Zuccolo A, Yu Y, Song X, Piegu B, Chevalier F, Walling JG, Ma J, Talag J, Brar DS, SanMiguel PJ, Jiang N, Jackson SA, Panaud O, Wing RA (2007) Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza.The Plant Journal, 52, 342-351. |
| [8] | Bennetzen JL, Kellogg EA (1997) Do plants have a one-way ticket to genomic obesity?The Plant Cell, 9, 1509-1514. |
| [9] | Brar DS, Khush GS (1997) Alien introgression in rice.Plant Molecular Biology, 35, 35-47. |
| [10] | Chaudhary RC, Khush GS (1990) Breeding rice varieties for resistance against Chilo spp. of stem borers in Asia and Africa.International Journal of Tropical Insect Science, 11, 659-669. |
| [11] | Chen J, Bughio H, Chen DZ, Liu GJ, Zhang KL, Zhang JY (2006) Development of chromosomal segment substitution lines from a backcross recombinant inbred population of interspecific rice cross.Rice Science, 13, 15-21. |
| [12] | Chen J, Huang DR, Wang L, Liu GJ, Zhuang JY (2010) Identification of quantitative trait loci for resistance to whitebacked planthopper, Sogatella furcifera, from an interspecific cross Oryza sativa × O. rufipogon.Breed Science, 60, 153-159. |
| [13] | Chen JF, Huang QF, Gao DY, Wang JY, Lang YS, Liu TY, Li B, Bai ZT, Luis Goicoechea J, Liang CZ, Chen CB, Zhang WL, Sun SH, Liao Y, Zhang XM, Yang L, Song CL, Wang MJ, Shi JF, Liu G, Liu JJ, Zhou HL, Zhou WL, Yu QL, An N, Chen Y, Cai QL, Wang B, Liu BH, Min JM, Huang Y, Wu HL, Li ZY, Zhang YQ, Yin Y, Song WQ, Jiang JM, Jackson SA, Wing RA, Wang J, Chen MS (2013) Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution.Nature Communications, 4, doi:10.1038/ncomms2596 |
| [14] | Chen XR, Yang KS, Fu JR, Zhu CI, Peng XS, He XP, He HH (2008) Identification and genetic analysis of fertility restoration ability in Dongxiang wild rice (Oryza rufipogon).Rice Science, 15, 21-28. |
| [15] | Chen ZW, Hu FY, Xu P, Li J, Deng XN, Zhou JW, Li F, Chan SN, Tao DY (2009) QTL analysis for hybrid sterility and plant height in interspecific populations derived from a wild rice relative, Oryza longistaminata.Breed Science, 59, 441-445. |
| [16] | Cheng ZK, Buell CR, Wing RA, Gu MH, Jiang JM (2001) Toward a cytological characterization of the rice genome.Genome Research, 11, 2133-2141. |
| [17] | Cheng ZK, Dong FG, Langdon T, Ouyang S, Buell CR, Gu MH, Blattner FR, Jiang JM (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon.The Plant Cell, 14, 1691-1704. |
| [18] | Chu YE, Oka HI (1970a) The genetic basis of crossing barriers between Oryza perennis subsp. barthii and its related taxa.Evolution, 24, 135-144. |
| [19] | Chu YE, Oka HI (1970b) Introgression across isolating barriers in wild and cultivated Oryza species.Evolution, 24, 344-355. |
| [20] | Dally AM, Second G (1990) Chloroplast DNA diversity in wild and cultivated species of rice (Genus Oryza, section Oryza). Cladistic-mutation and genetic-distance analysis.Theoretical and Applied Genetics, 80, 209-222. |
| [21] | Devadath S (1983) A strain of Oryza barthii, an African wild rice immune to bacterial blight of rice.Current Science, 52, 27-28. |
| [22] | Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication.Cell, 127, 1309-1321. |
| [23] | Fan CZ, Walling JG, Zhang JW, Hirsch CD, Jiang JM, Wing RA (2011) Conservation and purifying selection of transcribed genes located in a rice centromere.The Plant Cell, 23, 2821-2830. |
| [24] | Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics.Nature Reviews Genetics, 3, 329-341 |
| [25] | Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW (2010) Direct detection of DNA methylation during single-molecule, real-time sequencing.Nature Methods, 7, 461-465. |
| [26] | Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proceedings of the National Academy of Sciences,USA, 97, 4718-4723. |
| [27] | Fu Q, Zhang PJ, Tan LB, Zhu ZF, Ma D, Fu YC, Zhan XC, Cai HW, Sun CQ (2010) Analysis of QTLs for yield-related traits in Yuanjiang common wild rice (Oryza rufipogon Griff.).Journal of Genetics and Genomics, 37, 147-157. |
| [28] | Fu XL, Lu YG, Liu XD, Li JQ (2008) Progress on transferring elite genes from non-AA genome wild rice into Oryza sativa through interspecific hybridization.Rice Science, 15, 79-87. |
| [29] | Furuya A, Itoh R, Ishii R (1994) Mechanisms of different responses of leaf photosynthesis in African rice (Oryza glaberrima Steud.) and rice (Oryza sativa L.) to low leaf water potential.Japanese Journal of Crop Science, 63, 625-631. |
| [30] | Ge S, Sang T, Lu BR, Hong DY (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proceedings of the National Academy of Sciences,USA, 96, 14400-14405. |
| [31] | Goicoechea JL, Ammiraju JSS, Marri PR, Chen MS, Jackson S, Yu Y, Rounsley S, Wing RA (2010) The future of rice genomics: sequencing the collective Oryza genome.Rice, 3, 89-97. |
| [32] | Gutiérrez AG, Carabali SJ, Giraldo OX, Martinez CP, Correa F, Prado G, Tohme J, Lorieux M (2010) Identification of a rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × O. glaberrima introgression lines.BMC Plant Biology, 10, 6. |
| [33] | Haddrill PR, Halligan DL, Tomaras D, Charlesworth B (2007) Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over.Genome Biology, 8, R18. |
| [34] | Harland JR, De-Wet MJ (1971) Toward a rational classification of cultivated plants.Taxon, 20, 509-517. |
| [35] | Hass-Jacobus BL, Futrell-Griggs M, Abernathy B, Westerman R, Goicoechea JL, Stein J, Klein P, Hurwitz B, Zhou B, Rakhshan F, Sanyal A, Gill N, Lin JY, Walling JG, Luo MZ, Ammiraju JS, Kudrna D, Kim HR, Ware D, Wing RA, San Miguel P, Jackson SA (2006) Integration of hybridization-based markers (overgos) into physical maps for comparative and evolutionary explorations in the genus Oryza and in Sorghum.BMC Genomics, 7, 199. |
| [36] | Hastie AR, Dong L, Smith A, Finklestein J, Lam ET, Huo N, Cao H, Kwok PY, Deal KR, Dvorak J, Luo MC, Gu Y, Xiao M (2013) Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome.PLoS ONE, 8, e55864. |
| [37] | Heinrichs EA, Viajante VD, Romena AM (1985) Resistance of wild rices, Oryza spp. to the whorl maggot Hydrellia- Philippina Ferino (Diptera, Ephydridae).Environmental Entomology, 14, 404-407. |
| [38] | Heitz E (1928) Das Heterochromatin der Moose, pp. 762-818. Jahrb Wiss Botanik. Publishers , Bornträger. |
| [39] | Heuer S, Miézan KM (2003) Assessing hybrid sterility in Oryza glaberrima × O. sativa hybrid progenies by PCR marker analysis and crossing with wide compatibility varieties.Theoretical and Applied Genetics, 107, 902-909. |
| [40] | Hoan NT, Sarma NP, Siddiq EA (1997) Identification and characterization of new sources of cytoplasmic male sterility in rice.Plant Breeding, 116, 547-551. |
| [41] | Huisinga KL, Brower-Toland B, Elgin SCR (2006) The contradictory definitions of heterochromatin: transcription and silencing.Chromosoma, 115, 110-122. |
| [42] | International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome.Nature, 436, 793-800. |
| [43] | Jacquemin J, Chaparro C, Laudie M, Berger A, Gavory F, Goicoechea JL, Wing RA, Cooke R (2011) Long-range and targeted ectopic recombination between the two homeologous chromosomes 11 and 12 in Oryza species.Molecular Biology and Evolution, 28, 3139-3150. |
| [44] | Jacquemin J, Laudié M, Cooke R (2009) A recent duplication revisited: phylogenetic analysis reveals an ancestral duplication highly-conserved throughout the Oryza genus and beyond.BMC Plant Biology, 9, 146. |
| [45] | Jena KK, Khush GS (1990) Introgression of genes from Oryza officinalis Well ex Watt to cultivated rice, O. sativa L. Theoretical and Applied Genetics, 80, 737-745. |
| [46] | Jiao YL, Jia PX, Wang XF, Su N, Yu SL, Zhang DF, Ma LG, Feng Q, Jin ZQ, Li L, Xue YB, Cheng ZK, Zhao HY, Han B, Deng XW (2005) A tiling microarray expression analysis of rice chromosome 4 suggests a chromosome-level regulation of transcription.The Plant Cell, 17, 1641-1657. |
| [47] | Jin FX, Kwon SJ, Kang KH, Jeong OY, Le LH, Yon DB, Ahn SN (2004) Introgression for grain traits from Oryza minuta into rice, O. sativa.Rice Genetics Newsletter, 21, 15. |
| [48] | Khush GS (1997) Origin, dispersal, cultivation and variation of rice.Plant Molecular Biology, 35, 25-34. |
| [49] | Kim H, Hurwitz B, Yu Y, Collura K, Gill N, SanMiguel P, Mullikin JC, Maher C, Nelson W, Wissotski M, Braidotti M, Kudrna D, Goicoechea JL, Stein L, Ware D, Jackson SA, Soderlund C, Wing RA (2008) Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza.Genome Biology, 9, R45. |
| [50] | Kobayashi N, Ikeda R, Domingo IT, Vaughan DA (1993) Resistance to infection of rice tungro viruses and vector resistance in wild species of rice (Oryza spp.).Japanese Journal of Breeding, 43, 377-387. |
| [51] | Kobayashi N, Ikeda R, Vaughan DA (1994) Screening wild species of rice (Oryza spp.) for resistance to rice tungro disease.Japan Agricultural Research Quarterly, 28, 230-236. |
| [52] | Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, Kwok PY (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly.Nature Biotechnology, 30, 771-776. |
| [53] | Li DJ, Sun CQ, Fu YC, Li C, Zhu ZF, Chen L, Cai HW, Wang XK (2002) Identification and mapping of genes for improving yield from Chinese common wild rice (O. rufipogon Griff.) using advanced backcross QTL analysis.Chinese Science Bulletin, 47, 1533-1537. |
| [54] | Linh LH, Jin FS, Kang KH, Lee YT, Kwon SJ, Ahn SN (2006) Mapping quantitative trait loci for heading date and awn length using an advanced backcross line from a cross between Oryza sativa and O. minuta.Breeding Science, 56, 341-349. |
| [55] | Liu G, Lu G, Zeng L, Wang GL (2002) Two broad-spectrum blast resistance genes, Pi9(t) and Pi2(t), are physically linked on rice chromosome 6.Molecular Genetics and Genomics, 267, 472-480. |
| [56] | Lu BR, Sharma SD, Shastry SVS (1999) Taxonomy of the genus Oryza (Poaceae): historical perspective and current status.International Rice Research Newsletter, 24, 4-8. |
| [57] | Lu F, Ammiraju JSS, Sanyal A, Zhang SL, Song RT, Chen JF, Li GS, Sui Y, Song X, Cheng ZK, de Oliveira AC, Bennetzen JL, Jackson SA, Wing RA, Chen MS (2009) Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proceedings of the National Academy of Sciences,USA, 106, 2071-2076. |
| [58] | Ma JX, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice.Genome Research, 14, 860-869. |
| [59] | Martens JHA, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome.The EMBO Journal, 24, 800-812. |
| [60] | McCouch SR, Sweeney M, Li JM, Jiang H, Thomson M, Septiningsih E, Edwards J, Moncada P, Xiao JH, Garris A, Tai T, Martinez C, Tohme J, Sugiono M, McClung A, Yuan LP, Ahn SN (2007) Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa.Euphytica, 154, 317-339. |
| [61] | Miura K, Yamamota E, Morinaka Y, Takashi T, Kitano H, Matsuoka M, Ashikari M (2008) The hybrid breakdown 1(t) locus induces interspecific hybrid breakdown between rice Oryza sativa cv. Koshihikari and its wild relative O. nivara.Breeding Science, 58, 99-105. |
| [62] | Multani DS, Jena KK, Brar DS, de los Reyes BG, Angeles ER, Khush GS (1994) Development of monosomic alien addition lines and introgression of genes from Oryza australiensis Domin. to cultivated rice O. sativa L.Theoretical and Applied Genetics, 88, 102-109. |
| [63] | Nagaki K, Cheng ZK, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang JM (2004) Sequencing of a rice centromere uncovers active genes.Nature Genetics, 36, 138-145. |
| [64] | Nagaki K, Neumann P, Zhang DF, Ouyang S, Buell CR, Cheng ZK, Jiang JM (2005) Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice.Molecular Biology and Evolution, 22, 845-855. |
| [65] | Nanda JS (2000) Rice Breeding and Genetics: Research Priorities and Challenges. Science Publishers, Enfield. |
| [66] | Nanda JS, Sharma SD (2003) Monograph on Genus Oryza. Science Publishers, U.S. |
| [67] | Nayar NM (1968) Prevalence of self-incompatibility in Oryza barthii Cheval: its bearing on the evolution of rice and related taxa.Genetica, 38, 521-527. |
| [68] | Panaud O (2009) The molecular bases of cereal domestication and the history of rice.Comptes Rendus Biologies, 332, 267-272. |
| [69] | Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proceedings of the National Academy of Sciences,USA, 101, 9903-9908. |
| [70] | Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang HB, Wang XY, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang LF, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman R, Ware D, Westhoff P, Mayer KF, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses.Nature, 457, 551-556. |
| [71] | Piegu B, Guyot R, Picault N, Roulin A, Sanyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, Panaud O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice.Genome Research, 16, 1262-1269. |
| [72] | Prasad B, Eizenga GC (2008) Rice sheath blight disease resistance identified in Oryza spp. accessions.Plant Disease, 92, 1503-1509. |
| [73] | Rahman ML, Chu SH, Choi MS, Qiao YL, Jiang WZ, Piao RH, Khanam S, Cho YI, Jeung JU, Jena K, Koh HJ (2007) Identification of QTLs for some agronomic traits in rice using an introgression line from Oryza minuta.Molecules and Cells, 24, 16-26. |
| [74] | Ram T, Deen R, Gautam SK, Ramesh K, Rao YK, Brar DS (2010a) Identification of new genes for brown planthopper resistance in rice introgressed from O. glaberrima and O. minuta.Rice Genetics Newsletter, 25, 67. |
| [75] | Ram T, Laha GS, Gautam SK, Deen R, Madhav MS, Brar DS, Viraktamath BC (2010b) Identification of a new gene introgressed from Oryza brachyantha with broad-spectrum resistance to bacterial blight of rice in India.Rice Genetics Newsletter, 25, 57. |
| [76] | Ram T, Majumder ND, Padmavathi G, Mishra B (2005) Improving rice for broad-spectrum resistance to blast and salinity tolerance by introgressing genes from O. rufipogon.International Rice Research Notes, 30, 17-19. |
| [77] | Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Muller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C (2011) Comparative functional genomics of the fission yeasts.Science, 332, 930-936. |
| [78] | Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma LJ, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, Carlson JW, Carr A, Jungreis I, Marbach D, Sealfon R, Tolstorukov MY, Will S, Alekseyenko AA, Artieri C, Booth BW, Brooks AN, Dai Q, Davis CA, Duff MO, Feng X, Gorchakov AA, Gu T, Henikoff JG, Kapranov P, Li R, MacAlpine HK, Malone J, Minoda A, Nordman J, Okamura K, Perry M, Powell SK, Riddle NC, Sakai A, Samsonova A, Sandler JE, Schwartz YB, Sher N, Spokony R, Sturgill D, van Baren M, Wan KH, Yang L, Yu C, Feingold E, Good P, Guyer M, Lowdon R, Ahmad K, Andrews J, Berger B, Brenner SE, Brent MR, Cherbas L, Elgin SC, Gingeras TR, Grossman R, Hoskins RA, Kaufman TC, Kent W, Kuroda MI, Orr-Weaver T, Perrimon N, Pirrotta V, Posakony JW, Ren B, Russell S, Cherbas P, Graveley BR, Lewis S, Micklem G, Oliver B, Park PJ, Celniker SE, Henikoff S, Karpen GH, Lai EC, MacAlpine DM, Stein LD, White KP, Kellis M (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE.Science, 330, 1787-1797. |
| [79] | Sakamoto W, Kadowaki K, Tano S, Yabuno T (1990) Analysis of mitochondrial DNAs from Oryza glaberrima and its cytoplasmic substituted line for Oryza sativa associated with cytoplasmic male sterility.Japanese Journal of Genetics, 65, 1-6. |
| [80] | Sanyal A, Ammiraju JS, Lu F, Yu Y, Rambo T, Currie J, Kollura K, Kim HR, Chen JF, Ma XJ, San Miguel P, Chen MS, Wing RA, Jackson SA (2010) Orthologous comparisons of the Hd1 region across genera reveal Hd1 gene lability within diploid Oryza species and disruptions to microsynteny in Sorghum.Molecular Biology and Evolution, 27, 2487-2506. |
| [81] | Sarkar R, Raina SN (1992) Assessment of genome relationships in the genus Oryza L. based on seed-protein profile analysis.Theoretical and Applied Genetics, 85, 127-132. |
| [82] | Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du FY, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen WZ, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M, Kohlberg S, Sgro J, Delgado B, Mead K, Chinwalla A, Leonard S, Crouse K, Collura K, Kudrna D, Currie J, He R, Angelova A, Rajasekar S, Mueller T, Lomeli R, Scara G, Ko A, Delaney K, Wissotski M, Lopez G, Campos D, Braidotti M, Ashley E, Golser W, Kim H, Lee S, Lin J, Dujmic Z, Kim W, Talag J, Zuccolo A, Fan C, Sebastian A, Kramer M, Spiegel L, Nascimento L, Zutavern T, Miller B, Ambroise C, Muller S, Spooner W, Narechania A, Ren L, Wei S, Kumari S, Faga B, Levy MJ, McMahan L, Van Buren P, Vaughn MW, Ying K, Yeh CT, Emrich SJ, Jia Y, Kalyanaraman A, Hsia AP, Barbazuk WB, Baucom RS, Brutnell TP, Carpita NC, Chaparro C, Chia JM, Deragon JM, Estill JC, Fu Y, Jeddeloh JA, Han Y, Lee H, Li P, Lisch DR, Liu S, Liu Z, Nagel DH, McCann MC, SanMiguel P, Myers AM, Nettleton D, Nguyen J, Penning BW, Ponnala L, Schneider KL, Schwartz DC, Sharma A, Soderlund C, Springer NM, Sun Q, Wang H, Waterman M, Westerman R, Wolfgruber TK, Yang LX, Yu Y, Zhang LF, Zhou SG, Zhu QH, Bennetzen JL, Dawe RK, Jiang JM, Jiang N, Presting GG, Wessler SR, Aluru S, Martienssen RA, Clifton SW, McCombie WR, Wing RA, Wilson RK (2009) The B73 maize genome: complexity, diversity, and dynamics.Science, 326, 1112-1115. |
| [83] | Soderlund C, Nelson W, Shoemaker A, Paterson A (2006) SyMAP: a system for discovering and viewing syntenic regions of FPC maps.Genome Research, 16, 1159-1168. |
| [84] | Springer NM, Stupar RM (2007) Allelic variation and heterosis in maize: how do two halves make more than a whole?Genome Research, 17, 264-275. |
| [85] | Suh JP, Roh JH, Cho YC, Han SS, Kim YG, Jena KK (2009) The pi40 gene for durable resistance to rice blast and molecular analysis of pi40-advanced backcross breeding lines.Phytopathology, 99, 243-250. |
| [86] | Tang L, Zou XH, Achoundong G, Potgieter C, Second G, Zhang DY, Ge S (2010) Phylogeny and biogeography of the rice tribe (Oryzeae): evidence from combined analysis of 20 chloroplast fragments.Molecular Phylogenetics and Evolution, 54, 266-277. |
| [87] | Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild.Science, 277, 1063-1066. |
| [88] | Tian CC, Xiong YQ, Liu TY, Sun SH, Chen LB, Chen MS (2005) Evidence for an ancient whole-genome duplication event in rice and other cereals.Acta Genetica Sinica, 32, 519-527. |
| [89] | Tian ZX, Rizzon C, Du JC, Zhu LC, Bennetzen JL, Jackson SA, Gaut BS, Ma JX (2009) Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons?Genome Research, 19, 2221-2230. |
| [90] | Vales M (1985) Study of complete to Pyricularia oryzae Cav. of Oryza sativa × Oryza longistaminata hybrids and their Oryza longistaminata parent.Agronomie Tropicale, 40, 148-156. |
| [91] | Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus.Current Opinion in Plant Biology, 6, 139-146. |
| [92] | Wang XY, Shi XL, Hao BL, Ge S, Luo JC (2005) Duplication and DNA segmental loss in the rice genome: implications for diploidization.New Phytologist, 165, 937-946. |
| [93] | Wang XY, Tang HB, Bowers JE, Paterson AH (2009) Comparative inference of illegitimate recombination between rice and sorghum duplicated genes produced by polyploidization.Genome Research, 19, 1026-1032. |
| [94] | Wang YH, Li JY (2006) Genes controlling plant architecture.Current Opinion in Biotechnology, 17, 123-129. |
| [95] | Wang YH, Li JY (2008) Molecular basis of plant architecture.Annual Review of Plant Biology, 59, 253-279. |
| [96] | Wang ZY, Second G, Tanksley SD (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs.Theoretical and Applied Genetics, 83, 565-581. |
| [97] | Win KT, Kubo T, Miyazaki Y, Doi K, Yamagata Y, Yoshimura A (2009) Identification of two loci causing F1 pollen sterility in inter- and intraspecific crosses of rice.Breeding Science, 59, 411-418. |
| [98] | Wing RA, Ammiraju JS, Luo MZ, Kim H, Yu Y, Kudrna D, Goicoechea JL, Wang WM, Nelson W, Rao K, Brar D, Mackill DJ, Han B, Soderlund C, Stein L, SanMiguel P, Jackson S (2005) The Oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species.Plant Molecular Biology, 59, 53-62. |
| [99] | Wu JZ, Yamagata H, Hayashi-Tsugane M, Hijishita S, Fujisawa M, Shibata M, Ito Y, Nakamura M, Sakaguchi M, Yoshihara R, Kobayashi H, Ito K, Karasawa W, Yamamoto M, Saji S, Katagiri S, Kanamori H, Namiki N, Katayose Y, Matsumoto T, Sasaki T (2004) Composition and structure of the centromeric region of rice chromosome 8.The Plant Cell, 16, 967-976. |
| [100] | Wu YF, Kikuchi S, Yan HH, Zhang WL, Rosenbaum H, Iniguez AL, Jiang JM (2011) Euchromatic subdomains in rice centromeres are associated with genes and transcription.The Plant Cell, 23, 4054-4064. |
| [101] | Xing YZ, Zhang QF (2010) Genetic and molecular bases of rice yield.Annual Review of Plant Biology, 61, 421-442. |
| [102] | Xu X, Liu X, Ge S, Jensen JD, Hu FY, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, Li JX, He WM, Zhang GJ, Zheng XM, Zhang FM, Li YR, Yu C, Kristiansen K, Zhang XQ, Wang J, Wright M, McCouch S, Nielsen R, Wang J, Wang W (2011) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes.Nature Biotechnology, 30, 105-111. |
| [103] | Yang L, Liu TY, Li B, Sui Y, Chen JF, Shi JF, Wing RA, Chen MS (2012) Comparative sequence analysis of the Ghd7 orthologous regions revealed movement of Ghd7 in the grass genomes.PLoS ONE, 7, e50236. |
| [104] | Yu J, Wang J, Lin W, Li SG, Li H, Zhou J, Ni PX, Dong W, Hu SN, Zeng CQ, Zhang JG, Zhang Y, Li RQ, Xu ZY, Li XQ, Zheng HK, Cong LJ, Lin L, Yin JN, Geng JN, Li GY, Shi JP, Liu J, Lv H, Li J, Deng YJ, Ran LH, Shi XL, Wang XY, Wu QF, Li CF, Ren XY, Wang JQ, Wang XL, Li DW, Liu DY, Zhang XW, Ji ZD, Zhao WM, Sun YQ, Zhang ZP, Bao JY, Han YJ, Dong LL, Ji J, Chen P, Wu SM, Liu JS, Xiao Y, Bu DB, Tan JL, Yang L, Ye C, Zhang JF, Xu JY, Zhou Y, Yu YP, Zhang B, Zhuang SL, Wei HB, Liu B, Lei M, Yu H, Li YZ, Xu H, Wei SL, He XM, Fang LJ, Zhang ZJ, Zhang YZ, Huang XG, Su ZX, Tong W, Tong ZZ, Li SL, Ye J, Wang LS, Fang L, Lei TT, Chen C, Chen H, Xu Z, Li HH, Huang H, Zhang F, Xu HY, Li N, Zhao CF, Li ST, Dong LJ, Huang YQ, Li L, Xi Y, Qi QH, Li WJ, Hu W, Zhang YL, Tian XJ, Jiao YZ, Liang XH, Jin J, Gao L, Zheng WM, Hao BL, Liu SQ, Wang W, Yuan LP, Cao ML, McDermott J, Samudrala R, Wang J, Wong GK, Yang HM (2005) The genomes of Oryza sativa: a history of duplications.PLoS Biology, 3, e38. |
| [105] | Zamir D (2001) Improving plant breeding with exotic genetic libraries.Nature Reviews Genetics, 2, 983-989. |
| [106] | Zhu QH, Ge S (2005) Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes.New Phytologist, 167, 249-265. |
| [107] | Zou XH, Zhang FM, Zhang JG, Zang LL, Tang L, Wang J, Sang T, Ge S (2008) Analysis of 142 genes resolves the rapid diversification of the rice genus.Genome Biology, 9, R49. |
| [1] | QIU Fang, FU Jian-Min, JIN De-Min, WANG Bin. The molecular detection of genetic diversity [J]. Biodiv Sci, 1998, 06(2): 143-150. |
| [2] | LU Bao-Rong. Diversity of rice genetic resources and its utilization and conservation [J]. Biodiv Sci, 1998, 06(1): 63-72. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()
