



生物多样性 ›› 2011, Vol. 19 ›› Issue (3): 284-294. DOI: 10.3724/SP.J.1003.2011.11250 cstr: 32101.14.SP.J.1003.2011.11250
所属专题: 物种形成与系统进化
裴男才1,3,*( ), 张金龙2,3, 米湘成2, 葛学军1
), 张金龙2,3, 米湘成2, 葛学军1
收稿日期:2010-10-14
接受日期:2011-03-08
出版日期:2011-05-20
发布日期:2013-12-10
通讯作者:
裴男才
作者简介:*E-mail: nancai.pei@gmail.com基金资助:
Nancai Pei1,3,*( ), Jinlong Zhang2,3, Xiangcheng Mi2, Xuejun Ge1
), Jinlong Zhang2,3, Xiangcheng Mi2, Xuejun Ge1
Received:2010-10-14
Accepted:2011-03-08
Online:2011-05-20
Published:2013-12-10
Contact:
Nancai Pei
摘要:
系统发育群落生态学是近年兴起的一个重要生态学研究分支, 它以群落生态学为基础并引入了系统发育的分析方法, 全面动态地反映了群落中物种内和物种间的相互作用关系, 揭示了群落格局形成的生态学过程, 研究了生物多样性的形成及维持机制。巴拿马BCI (Barro Colorado Island)样地的成功例子说明, 在固定样地进行长期的群落生态与系统发育研究切实可行且极具意义; DNA条形码的快速兴起对这一研究发挥着重要作用。本文先列举了群落生态与系统发育综合分析能解决的群落系统发育结构、群落生态位结构、生物地理学和性状进化等生态学问题; 接着介绍了标准植物DNA条形码以及利用片段组合(rbcL+matK+trnH-psbA)进行快速物种识别和近缘种区分、精确群落系统发育关系的构建以及群落生态学研究; 随后提出DNA条形码研究在类群水平上需注意两片段的条形码组合(matK+rbcL)在同属种鉴别能力上的不足, 而在较大尺度群落水平上需对实验设计进行优化。DNA条形码将为探讨物种多样性及其维持机理、系统发育beta多样性以及群落水平上功能性状进化研究提供新的思路。
裴男才, 张金龙, 米湘成, 葛学军 (2011) 植物DNA条形码促进系统发育群落生态学发展. 生物多样性, 19, 284-294. DOI: 10.3724/SP.J.1003.2011.11250.
Nancai Pei, Jinlong Zhang, Xiangcheng Mi, Xuejun Ge (2011) Plant DNA barcodes promote the development of phylogenetic commu- nity ecology. Biodiversity Science, 19, 284-294. DOI: 10.3724/SP.J.1003.2011.11250.
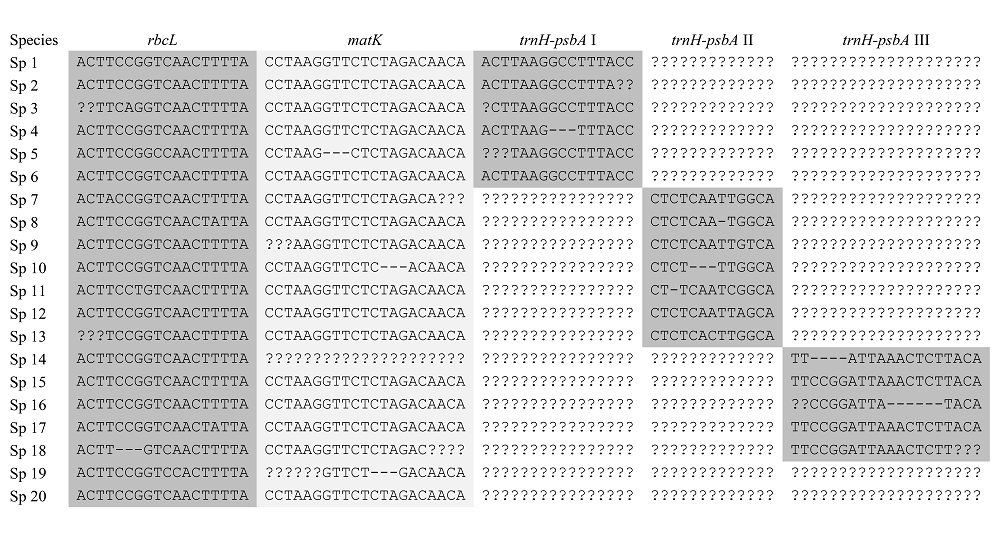
图1 rbcL+matK+trnH-psbA的3位点组合片段的超级矩阵示意图(改自Kress et al., 2009)。第一列为物种名称, 第二列之后的为各片段的序列排列情况, 其中trnH-psbA (也即trnH-psbA I, trnH-psbA II, trnH-psbA III等)片段以目为单位进行归类。3个片段共同组成一个矩阵。
Fig. 1 Illustration of supermatrix approach in sequence alignment for phylogenetic analysis (adopted from Kress et al., 2009). The first column represents species name, while others represent sequence alignments of rbcL, matK, and trnH-psbA. In trnH-psbA, each order consists of a dataset.
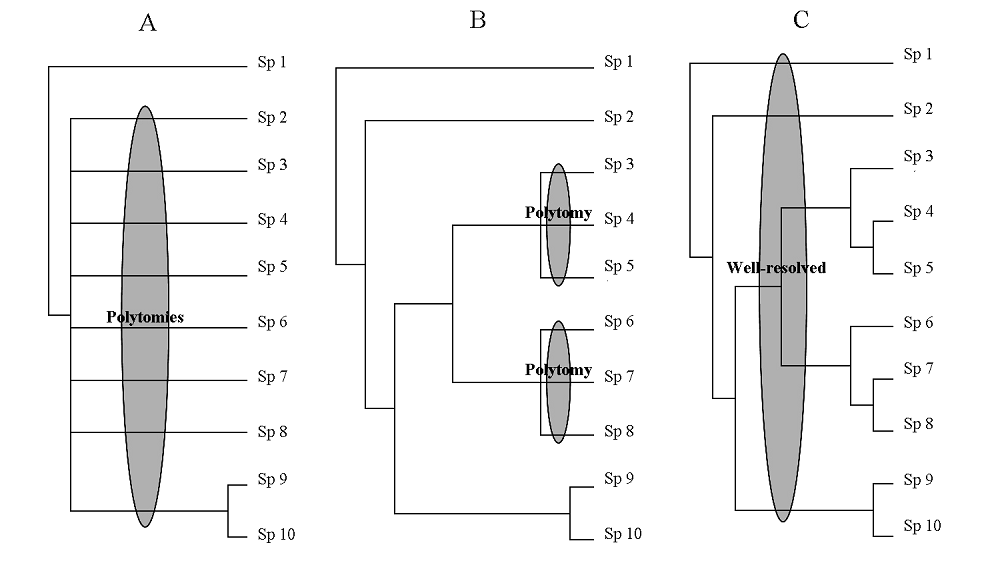
图2 10个物种的系统发育关系示意图。图A用Phylomatic的方法获得, 图B和C用条形码(Barcode)的方法获得。Sp 1-10表示物种1-10, 以此类推。图A表示当使用在线软件Phylomatic获取该10个物种的系统发育关系时, 物种2-10的位置未得到解决, 也就是常说的多分枝结构。图B表示, 当使用2个DNA条形码片段(如rbcL+matK)时, 物种3-5和物种6-8的系统发育关系尚不明确, 而其他物种的位置得到了较好解决。图C表示, 当使用3个DNA条形码片段构成的超级矩阵(如rbcL+matK+trnH-psbA)时, 全部物种的系统发育关系都得到了解决。
Fig. 2 A graphic representation of phylogenetic trees of ten species using Phylomatic (A) and Barcode (B, C) approaches. Sp 1-10 means species 1-10. (A) Phylogenetic relationships of Sp 2-10 are not well-resolved, i.e., there are polytomies among these taxa when using an online query tool Phylomatic; (B) Phylogenetic relationships of Sp 3-5, and Sp 6-8 are not well-resolved using a two-locus DNA barcode (rbcL+matK); (C) Phylogenetic relationships of Sp 1-10 are well-resolved using a supermatrix of three DNA barcodes (rbcL+matK+trnH-psbA).
| 标记 Marker | 引物 Primer | 序列 Sequence (5′- 3′) | 长度 Size (bp) | 参考文献 Reference |
|---|---|---|---|---|
| rbcL | SI_F | ATGTCACCACAAACAGAGACTAAAGC | 554 | |
| SI_R | GTYAAATCAAGTCCACCYCG | |||
| matK | KIM_3F | CGTACAGTACTTTTGTGTTTACGAG | Average 850 | Kim KJ, unpublished primers |
| KIM_1R | ACCCAGTCCATCTGGAAATCTTGGTTC | |||
| KIM_1F | AATATCCAAATACCAAATCC | Average 850 | Kim KJ, unpublished primers | |
| KIM_1R | ACCCAGTCCATCTGGAAATCTTGGTTC | |||
| 2.1a-f | ATCCATCTGGAAATCTTAGTTC | Average 890 | www.kew.org/barcoding/protocols.html | |
| 5r | GTTCTAGCACAAGAAAGTCG | |||
| trnH-psbA | psbA | GTTATGCATGAACGTAATGCTC | 280-750, average 470 | Sang et al., 1997 |
| trnH | CGCGCATGGTGGATTCACAATCC | Tate & Simpson (2003) |
附录I 本研究所使用的条形码片段引物对
Appendix 1 Primer pairs for barcode loci rbcL, matK, and trnH-psbA
| 标记 Marker | 引物 Primer | 序列 Sequence (5′- 3′) | 长度 Size (bp) | 参考文献 Reference |
|---|---|---|---|---|
| rbcL | SI_F | ATGTCACCACAAACAGAGACTAAAGC | 554 | |
| SI_R | GTYAAATCAAGTCCACCYCG | |||
| matK | KIM_3F | CGTACAGTACTTTTGTGTTTACGAG | Average 850 | Kim KJ, unpublished primers |
| KIM_1R | ACCCAGTCCATCTGGAAATCTTGGTTC | |||
| KIM_1F | AATATCCAAATACCAAATCC | Average 850 | Kim KJ, unpublished primers | |
| KIM_1R | ACCCAGTCCATCTGGAAATCTTGGTTC | |||
| 2.1a-f | ATCCATCTGGAAATCTTAGTTC | Average 890 | www.kew.org/barcoding/protocols.html | |
| 5r | GTTCTAGCACAAGAAAGTCG | |||
| trnH-psbA | psbA | GTTATGCATGAACGTAATGCTC | 280-750, average 470 | Sang et al., 1997 |
| trnH | CGCGCATGGTGGATTCACAATCC | Tate & Simpson (2003) |
| [1] |
Ackerly DD, Cornwell WK (2007) A trait-based approach to community assembly: partitioning of species trait values into within- and among-community components. Ecology Letters, 10, 135-145.
URL PMID |
| [2] |
Ackerly DD, Schwilk DW, Webb CO (2006) Niche evolution and adaptive radiation: testing the order of trait divergence. Ecology, 87, S50-S61.
DOI URL PMID |
| [3] | APG III (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG. III. Botanical Journal of the Linnean Society, 161, 105-121. |
| [4] | Baraloto C, Paine CET, Patino S, Bonal D, Herault B, Chave J (2009) Functional trait variation and sampling strategies in species-rich plant communities. Functional Ecology, 24, 208-216. |
| [5] | Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL (2008) Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proceedings of the National Academy of Sciences, USA, 105, 11505-11511. |
| [6] |
Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW (2009) The merging of community ecology and phylogenetic biology. Ecology Letters, 12, 693-715.
DOI URL PMID |
| [7] | CBOL Plant Working Group (2009) A DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA, 106, 12794-12797. |
| [8] | Chase JM, Leibold MA (2003) Ecological Niches: Linking Classical and Contemporary Approaches. University of Chicago Press, Chicago. |
| [9] | Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madrinan S, Petersen G, Seberg O, Jorgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barracl- ough TG, Kelly L, Wilkinson M (2007) A proposal for a standardised protocol to barcode all land plants. Taxon, 56, 295-299. |
| [10] |
Chen S, Yao H, Han J, Liu C, Song J, Shi L, Zhu Y, Ma X, Gao T, Pang X, Luo K, Li Y, Li X, Jia X, Lin Y, Leon C (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE, 5, e8613.
URL PMID |
| [11] | Chen SB (陈圣宾), Ouyang ZY (欧阳志云), Xu WH (徐卫华), Xiao Y (肖燚) (2010) A review of beta diversity studies. Biodiversity Science (生物多样性), 18, 323-335. (in Chinese with English abstract) |
| [12] | Condit R, Pitman N Jr, Leigh EG, Chave J, Terborgh J, Foster RB, Núñez P, Aguilar S, Valencia R, Villa G, Muller- Landau HC, Losos E, Hubbell SP (2002) Beta-diversity in tropical forest trees. Science, 295, 666-669. |
| [13] |
Cubas P (2004) Floral zygomorphy, the recurring evolution of a successful trait. Bioessays, 26, 1175-1184.
DOI URL PMID |
| [14] |
Dick CW, Kress WJ (2009) Dissecting tropical plant diversity with forest plots and a molecular toolkit. BioScience, 59, 745-755.
DOI URL |
| [15] |
Donoghue MJ (2008) A phylogenetic perspective on the distribution of plant diversity. Proceedings of the National Academy of Sciences, USA, 105, 11549-11555.
DOI URL |
| [16] | Eggleton P, Vane-Wright RI (1994) Phylogenetics and Ecology. Academic Press, London. |
| [17] |
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biological Conservation, 61, 1-10.
DOI URL |
| [18] |
Faith DP, Baker AM (2006) Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evolutionary Bioinformatics Online, 2, 121-128.
URL PMID |
| [19] | Faith DP, Ferrier S, Rosauer D (2010) Phylogenetic beta diversity as a tool in biodiversity conservation and monitor- ing. Powerpoint Presentation at Ecological Society of America Symposium. Pittsburgh, PA. |
| [20] |
Faith DP, Lozupone CA, Nipperess D, Knight R (2009) The cladistic basis for the phylogenetic diversity (PD) measure links evolutionary features to environmental gradients and supports broad applications of microbial ecology’s “phylogenetic beta diversity” framework. International Journal of Molecular Sciences, 10, 4723-4741.
DOI URL PMID |
| [21] | Fang JY (方精云) (2009) Community ecology comes to a new era. Biodiversity Science (生物多样性), 17, 531-532. (in Chinese) |
| [22] | Gómez JP, Bravo GA, Brumfield RT, Tello JG, Cadena CD (2010) A phylogenetic approach to disentangling the role of competition and habitat filtering in community assembly of Neotropical forest birds. Journal of Ecology, 79, 1181-1192. |
| [23] |
Gonzalez MA, Baraloto C, Engel J, Mori SA, Pétronelli P, Riéra B, Roger A, Thébaud C, Chave J (2009) Identification of Amazonian trees with DNA barcodes. PLoS ONE, 4, e7483.
URL PMID |
| [24] | Gonzalez MA, Roger A, Courtois EA, Jabot F, Norden N, Paine CET, Baraloto C, Thébaud C, Chave J (2010) Shifts in species and phylogenetic diversity between sapling and tree communities indicate negative density dependence in a lowland rain forest. Journal of Ecology, 98, 137-146. |
| [25] |
Graham CH, Fine PVA (2008) Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecology Letters, 11, 1265-1277.
DOI URL PMID |
| [26] |
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52, 696-704.
DOI URL PMID |
| [27] | Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences, USA, 103, 968-971. |
| [28] | Harvey PH, Pagel MD (1991) The Comparative Methods in Evolutionary Biology. Oxford University Press, Oxford. |
| [29] |
Hebert PDN, Ratnasingham S, de Waard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London Series B: Biological Sciences, 270, S96-S99.
DOI URL PMID |
| [30] |
Hewitt N (1998) Seed size and shade-tolerance: a comparative analysis of North American temperate trees. Oecologia, 114, 432-440.
URL PMID |
| [31] | Hubbell SP (2001) The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press, Princeton. |
| [32] |
Ives AR, Helmus MR (2010) Phylogenetic metrics of community similarity. The American Naturalist, 176, E128-E142.
DOI URL PMID |
| [33] |
Johnson MTJ, Stinchcombe JR (2007) An emerging synthesis between community ecology and evolutionary biology. Trends in Ecology and Evolution, 22, 250-257.
DOI URL PMID |
| [34] |
Kembel SW, Hubbell SP (2006) The phylogenetic structure of a neotropical forest tree community. Ecology, 87, S86-S99.
DOI URL PMID |
| [35] |
Kraft NJB, Cornwell WK, Webb CO, Ackerly DD (2007) Trait evolution, community assembly, and the phylogenetic structure of ecological communities. The American Naturalist, 170, 271-283.
DOI URL PMID |
| [36] | Kraft NJB, Metz MR, Condit RS, Chave J (2010) The relationship between wood density and mortality in a global tropical forest data set. New Phytologist, 188, 1124-1136. |
| [37] | Kraft NJB, Valencia R, Ackerly DD (2008) Functional traits and niche-based tree community assembly in an Amazonian forest. Science, 322, 580-582. |
| [38] |
Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE, 2, e508.
URL PMID |
| [39] | Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E (2009) Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Science, USA, 106, 18621-18626. |
| [40] |
Kress WJ, Erickson DL, Swenson NG, Thompson J, Uriarte M, Zimmerman JK (2010) Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PLoS ONE, 5, e15409.
DOI URL PMID |
| [41] | Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. Proceedings of the National Academy of Sciences, USA, 105, 2923-2928. |
| [42] | Lomáscolo SB, Levey DJ, Kimball RT, Bolker BM, Alborn HT (2010) Dispersers shape fruit diversity in Ficus (Moraceae). Proceedings of the National Academy of Sciences, USA, 107, 14668-14672. |
| [43] | Losos JB (2008) Phylogenetic niche conservatism, phylog- enetic signal and the relationship between phylogenetic rela- tedness and ecological similarity among species. Ecology Letters, 11, 995-1003. |
| [44] |
Lozupone C, Hamady M, Knight R (2006) UniFrac―an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics, 7, 371.
URL PMID |
| [45] | Ma KP (马克平) (2008) Large scale permanent plots: important platform for long term research on biodiversity in forest ecosystem. Journal of Plant Ecology (Chinese version) (植物生态学报), 32, 237. (in Chinese) |
| [46] |
Ma KP (马克平) (2011) Assessing progress of biodiversity conservasion with monitoring approach. Biodiversity Science (生物多样性), 19, 125-126. (in Chinese)
DOI URL |
| [47] |
Mayfield MM, Boni MF, Ackerly DD (2009) Traits, habitats, and clades: identifying traits of potential importance to environmental filtering. The American Naturalist, 174, E1-E22.
DOI URL PMID |
| [48] |
Niinemets Ü, Valladares F (2006) Tolerance to shade, drought, and waterlogging of temperate northern hemisphere trees and shrubs. Ecological Monographs, 76, 521-547.
DOI URL |
| [49] |
Ning SP (宁淑萍), Yan HF (颜海飞), Hao G (郝刚), Ge XJ (葛学军) (2008) Current advances of DNA barcoding study in plants. Biodiversity Science (生物多样性), 16, 417-425. (in Chinese with English abstract)
DOI URL |
| [50] |
Niu KC (牛克昌), Liu YN (刘怿宁), Shen ZH (沈泽昊), He FL (何芳良), Fang JY (方精云) (2009) Community assembly: the relative importance of neutral theory and niche theory. Biodiversity Science (生物多样性), 17, 579-593. (in Chinese with English abstract)
DOI URL |
| [51] |
Pavoine S, Vela E, Gachet S, Bélair GD, Bonsall MB (2011) Linking patterns in phylogeny, traits, abiotic variables and space: a novel approach to linking environmental filtering and plant community assembly. Journal of Ecology, 99, 165-175
DOI URL |
| [52] |
Pennisi E (2007) Wanted: a barcode for plants. Science, 318, 190-191.
URL PMID |
| [53] |
Rao CR (1982) Diversity and dissimilarity coefficients: a unified approach. Theoretical Population Biology, 21, 24-43.
DOI URL |
| [54] |
Ren B-Q, Xiang X-G, Chen Z-D (2010) Species identification of Alnus (Betulaceae) using nrDNA and cpDNA genetic markers. Molecular Ecology Resources, 10, 594-605.
URL PMID |
| [55] | Roy S, Tyagi A, Shukla V, Kumar A, Singh UM, Chaudhary LB, Datt B, Bag SK, Singh PK, Nair NK, Husain T, Tuli R (2010) Universal plant DNA barcode loci may not work in complex groups: a case study with Indian Berberis species. PLoS ONE, 5, e13674. |
| [56] |
Sanderson MJ (2003) r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics, 19, 301-302.
URL PMID |
| [57] | Silvertown J (2004) Plant coexistence and the niche. Trends in Ecology and Evolution, 19, 605-611. |
| [58] |
Stamatakis A (2006) RA×ML-VI-HPC: maximum likelihood- based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688-2690.
DOI URL PMID |
| [59] |
Swenson NG, Enquist BJ (2007) Ecological and evolutionary determinants of a key plant functional trait: wood density and its community-wide variation across latitude and elevation. American Journal of Botany, 94, 451-459.
DOI URL PMID |
| [60] |
Swenson NG, Enquist BJ, Pither J, Thompson J, Zimmerman JK (2006) The problem and promise of scale dependency in community phylogenetics. Ecology, 87, 2418-2424.
DOI URL |
| [61] | Tilman D (1988) Plant Strategies and the Dynamics and Structure of Plant Communities. Princeton University Press, Princeton, NJ. |
| [62] | Valladares F, Niinemets Ü (2008) Shade tolerance, a key plant feature of complex nature and consequences. Annual Review of Ecology, Evolution, and Systematics, 39, 237-257. |
| [63] |
van de Wiel CCM, van der Schoot J, van Valkenburg JLCH, Duistermaat H, Smulders MJM (2009) DNA barcoding discriminates the noxious invasive plant species, floating pennymort (Hydrocotyle ranunculoides L. f.), from non-invasive relatives. Molecular Ecology Resources, 9, 1086-1091.
DOI URL PMID |
| [64] | Wang X-Q (汪小全), Hong D-Y (洪德元) (1997) Progress in molecular systematics of plants in recent five years. Acta Phytotaxonomica Sinica (植物分类学报), 35, 465-480. (in Chinese with English abstract) |
| [65] |
Webb CO (2000) Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. The American Naturalist, 156, 145-155.
DOI URL PMID |
| [66] | Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Annual Review of Ecology and Systematics, 33, 475-505. |
| [67] | Webb CO, Losos JB, Agrawal AA (2006) Integrating phylogenies into community ecology. Ecology, 87, S1-S2. |
| [68] | Whittaker RH (1960) Vegetation of the Siskiyou Mountains, Oregon and California. Ecological Monographs, 30, 279-338. |
| [69] |
Wiens JJ (2003) Missing data, incomplete taxa, and phylogenetic accuracy. Systematic Biology, 52, 528-538.
DOI URL PMID |
| [70] |
Wright IJ, Ackerly DD, Bongers F, Harms KE, Ibarra- Manriquez G, Martinez-Ramos M, Mazer SJ, Muller- Landau HC, Paz H, Pitman NCA, Poorter L, Silman MR, Vriesendorp CF, Webb CO, Westoby M, Wright SJ (2007) Relationships among ecologically important dimensions of plant trait variation in seven Neotropical forests. Annals of Botany, 99, 1003-1015.
DOI URL PMID |
| [71] |
Wright SJ, Kitajima K, Kraft N, Reich P, Wright I, Bunker D, Condit R, Dalling J, Davies S, Diaz S, Engelbrecht B, Harms K, Hubbell S, Marks C, Ruiz-Jaen M, Salvador C, Zanne A (2010) Functional traits and the growth-mortality tradeoff in tropical trees. Ecology, 91, 3664-3674.
DOI URL PMID |
| [72] | Zhong Y (钟扬), Shi SH (施苏华), Ren WW (任文伟) (2004) Molecular phylogeny analysis. In: Plant Life History Evolution and Reproductive Ecology (植物生活史与繁殖生态学) (ed. Zhang DY (张大勇)). Science Press, Beijing. (in Chinese) |
| [1] | 王彦平, 张敏楚, 詹成修. 嵌套分布格局研究进展: 分析方法、影响机制及保护应用[J]. 生物多样性, 2023, 31(12): 23314-. |
| [2] | 张健, 孔宏智, 黄晓磊, 傅声雷, 郭良栋, 郭庆华, 雷富民, 吕植, 周玉荣, 马克平. 中国生物多样性研究的30个核心问题[J]. 生物多样性, 2022, 30(10): 22609-. |
| [3] | 邹怡. 样本量不一致时的β多样性计算[J]. 生物多样性, 2021, 29(6): 790-797. |
| [4] | 牛红玉, 王峥峰, 练琚愉, 叶万辉, 沈浩. 群落构建研究的新进展: 进化和生态 相结合的群落谱系结构研究[J]. 生物多样性, 2011, 19(3): 275-283. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn