



Biodiv Sci ›› 2008, Vol. 16 ›› Issue (5): 498-502. DOI: 10.3724/SP.J.1003.2008.08074 cstr: 32101.14.SP.J.1003.2008.08074
• Original Papers • Previous Articles Next Articles
Zhijun Dong1,2,3, Hui Huang1,*( ), Liangmin Huang1, Yuanchao Li1, Xiubao Li1
), Liangmin Huang1, Yuanchao Li1, Xiubao Li1
Received:2008-04-02
Accepted:2008-05-21
Online:2008-09-20
Published:2008-09-20
Contact:
Hui Huang
Zhijun Dong, Hui Huang, Liangmin Huang, Yuanchao Li, Xiubao Li. PCR-RFLP analysis of large subunit rDNA of symbiotic dinoflagellates in scleractinian corals from Luhuitou fringing reef of Sanya, Hainan[J]. Biodiv Sci, 2008, 16(5): 498-502.
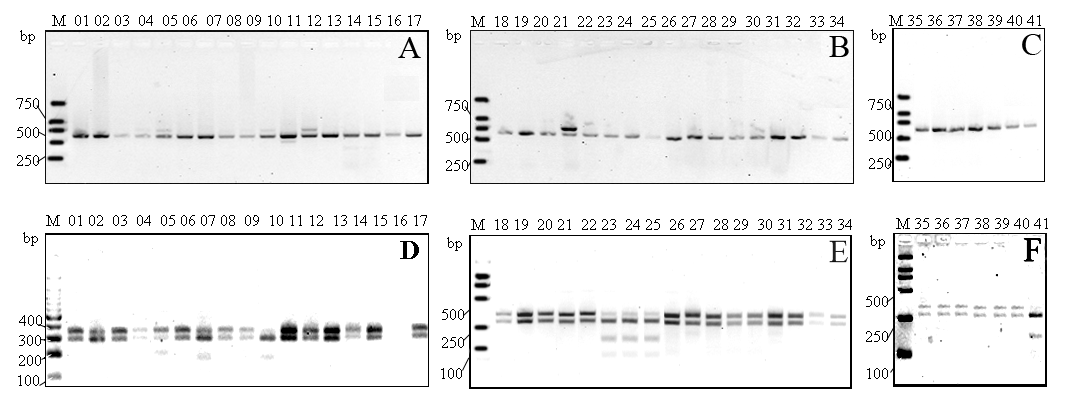
Fig. 1 Electrophoresis of the PCR-amplified 5' end of the 28S rDNA of Symbiodinium3(A, B, C) and RFLP genotypes of Symbiodiniumobtained from different colonies of scleractinian corals restricted by RsaI (D, E, F). M, DNA marker; 10, Pocillopora damicornis; 01, Acropora cerealis; 07, Acropora horrida; 15, Acropora cytherea; 18-20, 26, 28-30, 32, and 34, Acropora millepora; 14, Acropora sarmentosa; 06 and 17, Pavona decussata; 12, Pachyseris rugosa; 13, Pachyseris speciosa; 03, 21, 22, 27, 31, and 33, Porites lutea; 08, Porites compressa; 04, Porites murrayensis; 02, Goniopora gracilis; 38, Goniopora duofasciata; 23-25, Galaxea fascicularis; 35, Hydnophora exesa; 41, Favia favus; 11, Goniastrea pectinata; 16, Cyphastrea serailia; 39, Diploastrea heliopora; 37, Platygyra sinensis; 09, Plerogyra sinuosa.
| 珊瑚种类 Coral species | 采样数量 Sample size | 共生藻系群 Symbiodinium clade |
|---|---|---|
| 杯形珊瑚科 Pocilloporidae | ||
| 杯形珊瑚属 Pocillopora | ||
| 鹿角杯形珊瑚 P. damicornis | 1 | D |
| 鹿角珊瑚科 Acroporidae | ||
| 鹿角珊瑚属 Acropora | ||
| 丑鹿角珊瑚 A. horrida | 1 | C & D |
| 浪花鹿角珊瑚 A. cytherea | 1 | C |
| 多孔鹿角珊瑚 A. millepora | 9 | C |
| A. cerealis* | 1 | C |
| A. sarmentosa* | 1 | C |
| 菌珊瑚科 Agariciidae | ||
| 牡丹珊瑚属 Pavona | ||
| 十字牡丹珊瑚 P. decussata | 2 | C |
| 厚丝珊瑚属 Pachyseris | ||
| 皱纹厚丝珊瑚 P. rugosa | 1 | C |
| 标准厚丝珊瑚 P. speciosa | 1 | C |
| 滨珊瑚科 Poritidae | ||
| 滨珊瑚属 Porites | ||
| 澄黄滨珊瑚 P. lutea | 6 | C |
| 扁缩滨珊瑚 P. compressa | 1 | C |
| P. murrayensis* | 1 | C |
| 角孔珊瑚属 Goniopora | ||
| 细角孔珊瑚 G. gracilis | 1 | C |
| 二异角孔珊瑚 G. duofasciata | 1 | C |
| 枇杷珊瑚科 Oculinidae | ||
| 盔形珊瑚属 Galaxea | ||
| 丛生盔形珊瑚 G. fascicularis | 3 | C & D |
| 裸肋珊瑚科 Merulinidae | ||
| 刺柄珊瑚属 Hydnophora | ||
| 腐蚀刺柄珊瑚 H. exesa | 1 | C |
| 蜂巢珊瑚科 Faviidae | ||
| 蜂巢珊瑚属 Favia | ||
| 黄癣蜂巢珊瑚 F. favus | 1 | D |
| 菊花珊瑚属 Goniastrea | ||
| 梳状菊花珊瑚 G. pectinata | 1 | C |
| 刺星珊瑚属 Cyphastrea | ||
| 锯齿刺星珊瑚 C. serailia | 1 | C |
| 双星珊瑚属 Diploastrea | ||
| 同双星珊瑚 D. heliopora | 1 | C |
| 扁脑珊瑚属 Platygyra | ||
| 中华扁脑珊瑚 P. sinensis | 1 | C |
| 丁香珊瑚科 Caryophylliidae | ||
| 泡囊珊瑚属 Plerogyra | ||
| 泡囊珊瑚 P. sinuosa | 1 | C |
Table 1 Scleractinian coral species and their symbiotic dinoflagellates in Luhuitou fringing reef of Sanya
| 珊瑚种类 Coral species | 采样数量 Sample size | 共生藻系群 Symbiodinium clade |
|---|---|---|
| 杯形珊瑚科 Pocilloporidae | ||
| 杯形珊瑚属 Pocillopora | ||
| 鹿角杯形珊瑚 P. damicornis | 1 | D |
| 鹿角珊瑚科 Acroporidae | ||
| 鹿角珊瑚属 Acropora | ||
| 丑鹿角珊瑚 A. horrida | 1 | C & D |
| 浪花鹿角珊瑚 A. cytherea | 1 | C |
| 多孔鹿角珊瑚 A. millepora | 9 | C |
| A. cerealis* | 1 | C |
| A. sarmentosa* | 1 | C |
| 菌珊瑚科 Agariciidae | ||
| 牡丹珊瑚属 Pavona | ||
| 十字牡丹珊瑚 P. decussata | 2 | C |
| 厚丝珊瑚属 Pachyseris | ||
| 皱纹厚丝珊瑚 P. rugosa | 1 | C |
| 标准厚丝珊瑚 P. speciosa | 1 | C |
| 滨珊瑚科 Poritidae | ||
| 滨珊瑚属 Porites | ||
| 澄黄滨珊瑚 P. lutea | 6 | C |
| 扁缩滨珊瑚 P. compressa | 1 | C |
| P. murrayensis* | 1 | C |
| 角孔珊瑚属 Goniopora | ||
| 细角孔珊瑚 G. gracilis | 1 | C |
| 二异角孔珊瑚 G. duofasciata | 1 | C |
| 枇杷珊瑚科 Oculinidae | ||
| 盔形珊瑚属 Galaxea | ||
| 丛生盔形珊瑚 G. fascicularis | 3 | C & D |
| 裸肋珊瑚科 Merulinidae | ||
| 刺柄珊瑚属 Hydnophora | ||
| 腐蚀刺柄珊瑚 H. exesa | 1 | C |
| 蜂巢珊瑚科 Faviidae | ||
| 蜂巢珊瑚属 Favia | ||
| 黄癣蜂巢珊瑚 F. favus | 1 | D |
| 菊花珊瑚属 Goniastrea | ||
| 梳状菊花珊瑚 G. pectinata | 1 | C |
| 刺星珊瑚属 Cyphastrea | ||
| 锯齿刺星珊瑚 C. serailia | 1 | C |
| 双星珊瑚属 Diploastrea | ||
| 同双星珊瑚 D. heliopora | 1 | C |
| 扁脑珊瑚属 Platygyra | ||
| 中华扁脑珊瑚 P. sinensis | 1 | C |
| 丁香珊瑚科 Caryophylliidae | ||
| 泡囊珊瑚属 Plerogyra | ||
| 泡囊珊瑚 P. sinuosa | 1 | C |
| [1] |
Baker AC (2001) Reef corals bleach to survive change. Nature, 411,765-766.
DOI URL PMID |
| [2] | Baker AC (2003) Flexibility and specificity of coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annual Review of Ecology, Evolution, and Systematics, 34,661-689. |
| [3] |
Baker AC, Starger CJ, McClanahan TR, Glynn PW (2004) Corals’ adaptive response to climate change. Nature, 430,741.
URL PMID |
| [4] | Berkelmans R, van Oppen MJH(2006) The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proceedings of the Royal Society of London Series B: Biological Sciences, 273,2305-2312. |
| [5] | Carlos AA, Baillie BK, Kawachi M, Maruyama T (1999) Phylogenetic position of Symbiodinium (Dinophyceae) isolates from tridacnids (Bivalvia), cardiids (Bivalvia), a sponge (Porifera), a soft coral (Anthozoa), and a free living strain. Journal of Phycology, 35,1054-1062. |
| [6] | Chen CA, Lam KK, Nakano Y, Tsai WS (2003) A stable association of the stress-tolerant zooxanthellae, Symbiodinium clade D, with the low-temperature-tolerant coral, Oulastrea crispata (Scleractinia: Faviidae) in subtropical non-reefal coral communities. Zoological Studies, 42,540-550. |
| [7] | Chen CA, Wang JT, Fang LS, Yang YW (2005a) Fluctuating algal symbiont communities in Acropora palifera (Cnidaria; Scleractinia) from Taiwan. Marine Ecology Progress Series, 295,343-347. |
| [8] | Chen CA, Yang YW, Wei NV, Tsai WS, Fang LS (2005b) Symbiont diversity in scleractinian corals from tropical reefs and subtropical non-reef communities in Taiwan. Coral Reefs, 24,11-22. |
| [9] | Davies PS (1993) Endosymbiosis in marine cnidarians. In:,Plant-Animal Interactions in the Marine Benthos (eds John DM, Hawkins SJ, Price JH), pp. 511-540. Clarendon, Oxford. |
| [10] | Hoegh-Guldberg O (1999) Climate change, coral bleaching and the future of the world’s coral reefs. Marine Freshwater Research, 50,839-866. |
| [11] | Huang H, Dong ZJ, Huang LM, Zhang JB (2006) Restriction fragment length polymorphism analysis of large subunit rDNA of symbiotic dinoflagellates from scleractinian corals in the Zhubi coral reef of the Nansha Islands. Journal of Integrative Plant Biology, 48,148-152. |
| [12] |
Iglesias-Prieto R, Beltrán VH, LaJeunesse TC, Reyes-Bonilla H, Thome PE (2004) Different algal symbionts explain the vertical distribution of dominant reef corals in the eastern Pacific. Proceedings of the Royal Society of London Series B: Biological Sciences, 271,1757-1763.
DOI URL PMID |
| [13] | Kinzie RA III, Takayama M, Santos SR (2001) The adaptive bleaching hypothesis: experimental tests of critical assumptions. The Biological Bulletin, 200,51-58. |
| [14] |
LaJeunesse TC, Trench RK (2000) Biogeography of two species of Symbiodinium (Freudenthal) inhabiting the intertidal sea anemone Anthopleura elegantissima (Brandt). The Biological Bulletin, 199,126-134.
DOI URL PMID |
| [15] |
Lewis CL, Coffroth MA (2004) The acquisition of exogenous algal symbionts by an octocoral after bleaching. Science, 304,1490-1492.
URL PMID |
| [16] | Pochon X, Pawlowski J, Zaninetti L, Rowan R (2001) High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Marine Biology, 139,1069-1078. |
| [17] |
Pochon X, Montoya-Burgos JI, Stadelmann B, Pawlowski J (2006) Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Molecular Phylogenetics and Evolution, 38,20-30.
URL PMID |
| [18] |
Rodriguez-Lanetty M (2003) Evolving lineages of Symbiodinium-like dinoflagellates based on ITS1 rDNA. Molecular Phylogenetics and Evolution, 28,152-168.
DOI URL PMID |
| [19] | Rowan R (2004) Thermal adaptation in reef coral symbionts. Nature, 430,742. |
| [20] | Rowan R, Powers DA (1991) A molecular genetic classification of zooxanthellae and the evolution of animal-algal symbiosis. Science, 251,1348-1351. |
| [21] |
Rowan R, Knowlton N, Baker AC, Jara J (1997) Landscape ecology of algal symbiont communities explains variation in episodes of coral bleaching. Nature, 388,265-269.
DOI URL PMID |
| [22] | Toller WW, Rowan R, Knowlton N (2001a) Zooxanthellae of the Montastraea annularis species complex: patterns of distribution of four taxa of Symbiodinium on divergent reefs and across depths. The Biological Bulletin, 201,348-359. |
| [23] |
Toller WW, Rowan R, Knowlton N (2001b) Repopulation of zooxanthellae in the Caribbean corals Montastraea annularis and M. faveolata following experimental and disease associated bleaching. The Biological Bulletin, 201,360-373.
URL PMID |
| [24] | Yu DP (于登攀), Zou RL (邹仁林) (1996) Study on the species diversity of the scleractinian coral community on Luhuitou fringing reef. Acta Ecologica Sinica(生态学报), 16,469-475. (in Chinese with English abstract) |
| [25] | Zhang QM (张乔民), Shi Q (施祺), Chen G (陈刚), Fong TCW (方静威), Wong DCC (黄志俊), Huang H (黄晖), Wang HK (王汉奎), Zhao MX (赵美霞) (2006) Status monitoring and health assessment of Luhuitou fringing reef of Sanya, Hainan, China. Chinese Science Bulletin (科学通报), 51,81-88. (in Chinese) |
| [26] | Zou RL (邹仁林) (2001) Fauna Sinica: Hermatypic Coral(中国动物志‧造礁石珊瑚). Science Press,Beijing. (in Chinese) |
| [27] | Zou RL (邹仁林), Song SW (宋善文), Ma JH (马江虎) (1975) Shallow Water Hermatypic Coral in Hainan Island (海南岛浅水造礁石珊瑚). Science Press,Beijing. (in Chinese) |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()