



Biodiv Sci ›› 2014, Vol. 22 ›› Issue (3): 264-276. DOI: 10.3724/SP.J.1003.2014.14012 cstr: 32101.14.SP.J.1003.2014.14012
Special Issue: 生物多样性信息学专题(II)
• Biodiversity Informatics • Previous Articles Next Articles
Lisong Wang1,*( ), Vincent S Smith2, Hongrui Zhang1, Xianchun Zhang1
), Vincent S Smith2, Hongrui Zhang1, Xianchun Zhang1
Received:2014-01-13
Accepted:2014-04-30
Online:2014-05-20
Published:2014-06-04
Contact:
Wang Lisong
Lisong Wang, Vincent S Smith, Hongrui Zhang, Xianchun Zhang. Scratchpads 2.0: a virtual research environment for biodiversity sciences in the Internet era[J]. Biodiv Sci, 2014, 22(3): 264-276.
| 内容分类 Category | 站点名称 Name of sites | 使用的S2关键功能 Key Scratchpads features used |
|---|---|---|
| 农业和园艺 Agriculture and horticulture | 世界茄科在线 Solanaceae Source (http://solanaceae.myspecies.info/) | 植物分类 Botanical taxonomy 野外调查页面 Field work pages BRAHMS 数据导入 BRAHMS data import 物种多媒体 Taxonomic media 协同管理 Collaborator management |
| 动物多样性 Animal biodiversity | 非洲鱼类学门户 African Ichthyology Portal (http://africhthy.org/) | 多语言支持(英语/法语) Multi-lingual support (English/French) 社区论坛 Community forums 动物分类 Zoological taxonomy 学术文献 Scientific literature 物种多媒体 Taxonomic media |
| 公众科学 Citizen science | 硅藻在线 Diatoms Online (http://diatoms.myspecies.info/) | 达尔文核心标准 Darwin-core specimen records 动物分类 Zoological taxonomy 多用户博客 Multi-user blogs |
| 保护 Conservation | 植物红皮书索引 IUCN Sampled Red List Index for Plants (http://threatenedplants.myspecies.info/) | 保护评估 Conservation assessments IUCN数据整合 IUCN data integration 多元植物分类 Multiple botanical classifications 科学文献 Scientific literature |
| 入侵种 Invasive species | 认识蚂蚁 Antkey (http://antkey.org/) | 多语言支持(英语/中文/印度尼西亚语) Multi-lingual support (English/Chinese/ Indonesian) 解剖学词汇 Anatomical glossary 物种分布图 Species occurence maps |
| 植物多样性 Plant Biodiversity | 加拿大和阿拉斯加极地植物区系 Arctic Flora of Canada and Alaska (http://arcticplants.myspecies.info/) | 植物分类 Botanical taxonomy 自定义页面 Custom pages 自定义数据内容 Embedded custom content Google地图 Google Maps |
| 专业组织 Society | 国际蝽类昆虫学会 The International Heteropterists’ Society (http://ihs.myspecies.info/) | 个人群组 Private organisational groups 群组交流工具 Group communication tools 科学文献 Scientific literature 动物分类 Zoological taxonomy 物种多媒体 Taxonomic media |
| 系统学 Systematics | 蚊子的分类和编目 Mosquito Taxonomic Inventory (http://mosquito-taxonomic-inventory.info/) | 解剖学词汇 Anatomical glossary 动物分类 Zoological taxonomy 物种多媒体 Taxonomic media 图库 Galleries 科学文献 Scientific literature |
Table 1 Case study of using Scratchpads 2.0
| 内容分类 Category | 站点名称 Name of sites | 使用的S2关键功能 Key Scratchpads features used |
|---|---|---|
| 农业和园艺 Agriculture and horticulture | 世界茄科在线 Solanaceae Source (http://solanaceae.myspecies.info/) | 植物分类 Botanical taxonomy 野外调查页面 Field work pages BRAHMS 数据导入 BRAHMS data import 物种多媒体 Taxonomic media 协同管理 Collaborator management |
| 动物多样性 Animal biodiversity | 非洲鱼类学门户 African Ichthyology Portal (http://africhthy.org/) | 多语言支持(英语/法语) Multi-lingual support (English/French) 社区论坛 Community forums 动物分类 Zoological taxonomy 学术文献 Scientific literature 物种多媒体 Taxonomic media |
| 公众科学 Citizen science | 硅藻在线 Diatoms Online (http://diatoms.myspecies.info/) | 达尔文核心标准 Darwin-core specimen records 动物分类 Zoological taxonomy 多用户博客 Multi-user blogs |
| 保护 Conservation | 植物红皮书索引 IUCN Sampled Red List Index for Plants (http://threatenedplants.myspecies.info/) | 保护评估 Conservation assessments IUCN数据整合 IUCN data integration 多元植物分类 Multiple botanical classifications 科学文献 Scientific literature |
| 入侵种 Invasive species | 认识蚂蚁 Antkey (http://antkey.org/) | 多语言支持(英语/中文/印度尼西亚语) Multi-lingual support (English/Chinese/ Indonesian) 解剖学词汇 Anatomical glossary 物种分布图 Species occurence maps |
| 植物多样性 Plant Biodiversity | 加拿大和阿拉斯加极地植物区系 Arctic Flora of Canada and Alaska (http://arcticplants.myspecies.info/) | 植物分类 Botanical taxonomy 自定义页面 Custom pages 自定义数据内容 Embedded custom content Google地图 Google Maps |
| 专业组织 Society | 国际蝽类昆虫学会 The International Heteropterists’ Society (http://ihs.myspecies.info/) | 个人群组 Private organisational groups 群组交流工具 Group communication tools 科学文献 Scientific literature 动物分类 Zoological taxonomy 物种多媒体 Taxonomic media |
| 系统学 Systematics | 蚊子的分类和编目 Mosquito Taxonomic Inventory (http://mosquito-taxonomic-inventory.info/) | 解剖学词汇 Anatomical glossary 动物分类 Zoological taxonomy 物种多媒体 Taxonomic media 图库 Galleries 科学文献 Scientific literature |
| 数据和元数据标准 Data and metadata standards | 说明 Notes |
|---|---|
| BibTeX, RIS, XML bibliographic citations (export can also be in XML, RTF or Tagged Field) | 与各种桌面和在线文献管理工具, 如Endnote, ReferenceManager, BitText和Google Scholar等进行双向数据交换 |
| CSV / XLSX (spreadsheet content) | 与电子表格数据如Excel、Access等进行数据交换(需要使用指定的数据模板格式) |
| Darwin Core Archive and selected extensions | 达尔文数据标准, 标本数据交换标准 |
| EXIF, XMP image metadata | 图像元数据交换标准 |
| ITIS taxon metadata standard (for taxon names and hierarchies) | ITIS分类群元数据标准(适用于类群学名和分类树) |
| LUCID (for taxonomic keys) | 与鉴定工具LUCID系统的数据交换 |
| Nexus (character data, export only) | 与系统发育分析系统或性状矩阵软件的数据交换格式Nexus |
| Species Profile Model (for taxon descriptions) | TDWG推荐的结构化的物种描述模型标准 |
| XML (export only, selected content in the TaxPub schema, | NCBI所推荐的与NCBI相关期刊中物种描述的信息标准 |
Table 2 Data and metadata standards implemeneted in Scratchpads 2.0
| 数据和元数据标准 Data and metadata standards | 说明 Notes |
|---|---|
| BibTeX, RIS, XML bibliographic citations (export can also be in XML, RTF or Tagged Field) | 与各种桌面和在线文献管理工具, 如Endnote, ReferenceManager, BitText和Google Scholar等进行双向数据交换 |
| CSV / XLSX (spreadsheet content) | 与电子表格数据如Excel、Access等进行数据交换(需要使用指定的数据模板格式) |
| Darwin Core Archive and selected extensions | 达尔文数据标准, 标本数据交换标准 |
| EXIF, XMP image metadata | 图像元数据交换标准 |
| ITIS taxon metadata standard (for taxon names and hierarchies) | ITIS分类群元数据标准(适用于类群学名和分类树) |
| LUCID (for taxonomic keys) | 与鉴定工具LUCID系统的数据交换 |
| Nexus (character data, export only) | 与系统发育分析系统或性状矩阵软件的数据交换格式Nexus |
| Species Profile Model (for taxon descriptions) | TDWG推荐的结构化的物种描述模型标准 |
| XML (export only, selected content in the TaxPub schema, | NCBI所推荐的与NCBI相关期刊中物种描述的信息标准 |
| 序号 No. | 主题术语 Items | 解释和说明 Description |
|---|---|---|
| 1 | 关系 Associations | 描述捕食者-被捕食者; 寄主-寄生虫; 传粉, 共生, 互惠, 共栖, 杂交等生物相互关系的信息 Predator-prey; host-parasite, pollinators, symbiosis, mutualism, commensalism, hybridization… |
| 2 | 行为 Behaviour | 描述生物有机体对生物或非生物环境的反应 Cover actions and reactions of organism in relation to its biotic and abiotic environment |
| 3 | 保护状况 Conservation status | 描述物种目前或将来面临绝灭的可能性 A description of the likelihood of the species becoming extinct in the present day or in the near future |
| 4 | 周期性 Cyclicity | 描述物种周期性的特征, 比如物候 A state or condition characterised by regular repetition in time |
| 5 | 细胞学 Cytology | 描述物种的细胞生物学, 例如结构、功能等 Cell biology formation, structure and function of cells |
| 6 | 特征摘要 Diagnostic description | 描述物种与其相似物种的区别性特征 Distinguishing feature of this taxon from its closest relatives |
| 7 | 疾病 Diseases | 描述物种的疾病危害 Diseases of organisms |
| 8 | 散布 Dispersal | 描述物种的散布策略和机制 Dispersal strategies and mechanisms |
| 9 | 分布 Distribution | 描述物种按行政区域或生物地理区域界定的地理分布范围, 可以是全球性分布, 也可以是局部尺度上的分布情况。Cover ranges, e.g., a global range, or a narrower one; may be biogeographical, political or other (e.g., managed areas like conservencies); endemism; native or exotic; ref Darwin Core Geospatial extension. |
| 10 | 演化 Evolution | 描述物种的系统演化信息 Phylogenetic information relating to the taxon |
| 11 | 一般描述 General description | 广泛描述物种的综合信息 A comprehensive description of the characteristics of the taxon |
| 12 | 遗传学 Genetics | 有关物种的遗传信息, 包括染色体信息 Including karyotypes |
| 13 | 生长 Growth | 描述物种的生长速率、参数等 Rate; parameters; allometries. |
| 14 | 生境 Habitat | 描述物种的栖息环境, 包括地表分类(陆地、海洋)以及气候条件、忍耐幅、水平和垂直梯度上的特征 Include realm (e.g. Terrestrial etc) and climatic information (e.g. Boreal); also include requirements and tolerances; horizontal and vertical distribution. |
| 15 | 立法 Legislation | 描述有关该物种的立法情况 Legal regulations or statutes relating to the taxon |
| 16 | 生活周期 Life cycle | 专性发育转换 Obligatory developmental transformations |
| 17 | 寿命 Life expectancy | 描述物种的平均寿命 The average period an organism can be expected to survive |
| 18 | 相似物种 Look alikes | 描述与该物种相似的其他物种, 例如在入侵种群落中 Other taxa that this taxon may be confused with. Common in invasive species communities. |
| 19 | 管理 Management | 描述与物种的立法相关的管理情况, 比如CITES名录 A statement about the level of need to manage a taxon which can be related to a piece of legislation, e.g., a CITES list. |
| 20 | 迁移 Migration | 描述物种定期从一个地点移动到另外一个地点的情况, 例如动物繁殖期间的移动 Periodic movement of organisms from one locality to another (e.g., for breeding). |
| 21 | 分子生物学 Molecular biology | 描述物种的基因、蛋白质、生物化学(例如毒理) Include genomic, proteomic and biochemistry (e.g. toxicity) |
| 22 | 形态学 Morphology | 描述物种的形态特征(也包括解剖特征) The appearance of the taxon; e.g. habit; anatomy (the branch of morphology that deals with structure of animals) |
| 23 | 生理学 Physiology | 描述物种的生理学过程 An account of the physiological processes |
| 24 | 居群生物学 Population biology | 描述物种的丰度信息 Include abundance information |
| 25 | 管理步骤 Procedures | 如何处理某一类群, 已知的威胁是什么? Deal with how you go about managing this taxon; what are the known threats to this taxon? |
| 26 | 生殖 Reproduction | 描述物种的生殖策略、信号和限制条件 Reproduction cues, strategies, restraints. |
| 27 | 风险评估 Risk statement | 描述物种的入侵风险和影响 Include invasiveness, impact |
| 28 | 体积 Size | 描述物种的体积, 例如周长、长、体积、重量 Average size, max, range; type of size (perimeter, length, volume, weight ...) |
| 29 | 分类群生物学 Taxon biology | 描述分类群的生物学特征 An account of the biology of the taxon |
| 30 | 威胁 Threats | 描述物种面临的威胁 The threats to which this taxon is subject |
| 31 | 趋势 Trends | 描述物种的居群是否稳定?是在增长还是下降等信息 An indication of whether a population is stable, or increasing or decreasing. |
| 32 | 营养策略 Trophic strategy | 描述物种在食物网中的位置和食物偏好 Include nutritional aspects, diet, position in food network. |
| 33 | 用途 Uses | 描述物种与人类利用的关系: 参考“经济植物” Relationships to humans; ref Cook “Economic Botany” |
Table 3 Example standard of Species Profile Module implemeted in Scratchpads 2.0
| 序号 No. | 主题术语 Items | 解释和说明 Description |
|---|---|---|
| 1 | 关系 Associations | 描述捕食者-被捕食者; 寄主-寄生虫; 传粉, 共生, 互惠, 共栖, 杂交等生物相互关系的信息 Predator-prey; host-parasite, pollinators, symbiosis, mutualism, commensalism, hybridization… |
| 2 | 行为 Behaviour | 描述生物有机体对生物或非生物环境的反应 Cover actions and reactions of organism in relation to its biotic and abiotic environment |
| 3 | 保护状况 Conservation status | 描述物种目前或将来面临绝灭的可能性 A description of the likelihood of the species becoming extinct in the present day or in the near future |
| 4 | 周期性 Cyclicity | 描述物种周期性的特征, 比如物候 A state or condition characterised by regular repetition in time |
| 5 | 细胞学 Cytology | 描述物种的细胞生物学, 例如结构、功能等 Cell biology formation, structure and function of cells |
| 6 | 特征摘要 Diagnostic description | 描述物种与其相似物种的区别性特征 Distinguishing feature of this taxon from its closest relatives |
| 7 | 疾病 Diseases | 描述物种的疾病危害 Diseases of organisms |
| 8 | 散布 Dispersal | 描述物种的散布策略和机制 Dispersal strategies and mechanisms |
| 9 | 分布 Distribution | 描述物种按行政区域或生物地理区域界定的地理分布范围, 可以是全球性分布, 也可以是局部尺度上的分布情况。Cover ranges, e.g., a global range, or a narrower one; may be biogeographical, political or other (e.g., managed areas like conservencies); endemism; native or exotic; ref Darwin Core Geospatial extension. |
| 10 | 演化 Evolution | 描述物种的系统演化信息 Phylogenetic information relating to the taxon |
| 11 | 一般描述 General description | 广泛描述物种的综合信息 A comprehensive description of the characteristics of the taxon |
| 12 | 遗传学 Genetics | 有关物种的遗传信息, 包括染色体信息 Including karyotypes |
| 13 | 生长 Growth | 描述物种的生长速率、参数等 Rate; parameters; allometries. |
| 14 | 生境 Habitat | 描述物种的栖息环境, 包括地表分类(陆地、海洋)以及气候条件、忍耐幅、水平和垂直梯度上的特征 Include realm (e.g. Terrestrial etc) and climatic information (e.g. Boreal); also include requirements and tolerances; horizontal and vertical distribution. |
| 15 | 立法 Legislation | 描述有关该物种的立法情况 Legal regulations or statutes relating to the taxon |
| 16 | 生活周期 Life cycle | 专性发育转换 Obligatory developmental transformations |
| 17 | 寿命 Life expectancy | 描述物种的平均寿命 The average period an organism can be expected to survive |
| 18 | 相似物种 Look alikes | 描述与该物种相似的其他物种, 例如在入侵种群落中 Other taxa that this taxon may be confused with. Common in invasive species communities. |
| 19 | 管理 Management | 描述与物种的立法相关的管理情况, 比如CITES名录 A statement about the level of need to manage a taxon which can be related to a piece of legislation, e.g., a CITES list. |
| 20 | 迁移 Migration | 描述物种定期从一个地点移动到另外一个地点的情况, 例如动物繁殖期间的移动 Periodic movement of organisms from one locality to another (e.g., for breeding). |
| 21 | 分子生物学 Molecular biology | 描述物种的基因、蛋白质、生物化学(例如毒理) Include genomic, proteomic and biochemistry (e.g. toxicity) |
| 22 | 形态学 Morphology | 描述物种的形态特征(也包括解剖特征) The appearance of the taxon; e.g. habit; anatomy (the branch of morphology that deals with structure of animals) |
| 23 | 生理学 Physiology | 描述物种的生理学过程 An account of the physiological processes |
| 24 | 居群生物学 Population biology | 描述物种的丰度信息 Include abundance information |
| 25 | 管理步骤 Procedures | 如何处理某一类群, 已知的威胁是什么? Deal with how you go about managing this taxon; what are the known threats to this taxon? |
| 26 | 生殖 Reproduction | 描述物种的生殖策略、信号和限制条件 Reproduction cues, strategies, restraints. |
| 27 | 风险评估 Risk statement | 描述物种的入侵风险和影响 Include invasiveness, impact |
| 28 | 体积 Size | 描述物种的体积, 例如周长、长、体积、重量 Average size, max, range; type of size (perimeter, length, volume, weight ...) |
| 29 | 分类群生物学 Taxon biology | 描述分类群的生物学特征 An account of the biology of the taxon |
| 30 | 威胁 Threats | 描述物种面临的威胁 The threats to which this taxon is subject |
| 31 | 趋势 Trends | 描述物种的居群是否稳定?是在增长还是下降等信息 An indication of whether a population is stable, or increasing or decreasing. |
| 32 | 营养策略 Trophic strategy | 描述物种在食物网中的位置和食物偏好 Include nutritional aspects, diet, position in food network. |
| 33 | 用途 Uses | 描述物种与人类利用的关系: 参考“经济植物” Relationships to humans; ref Cook “Economic Botany” |
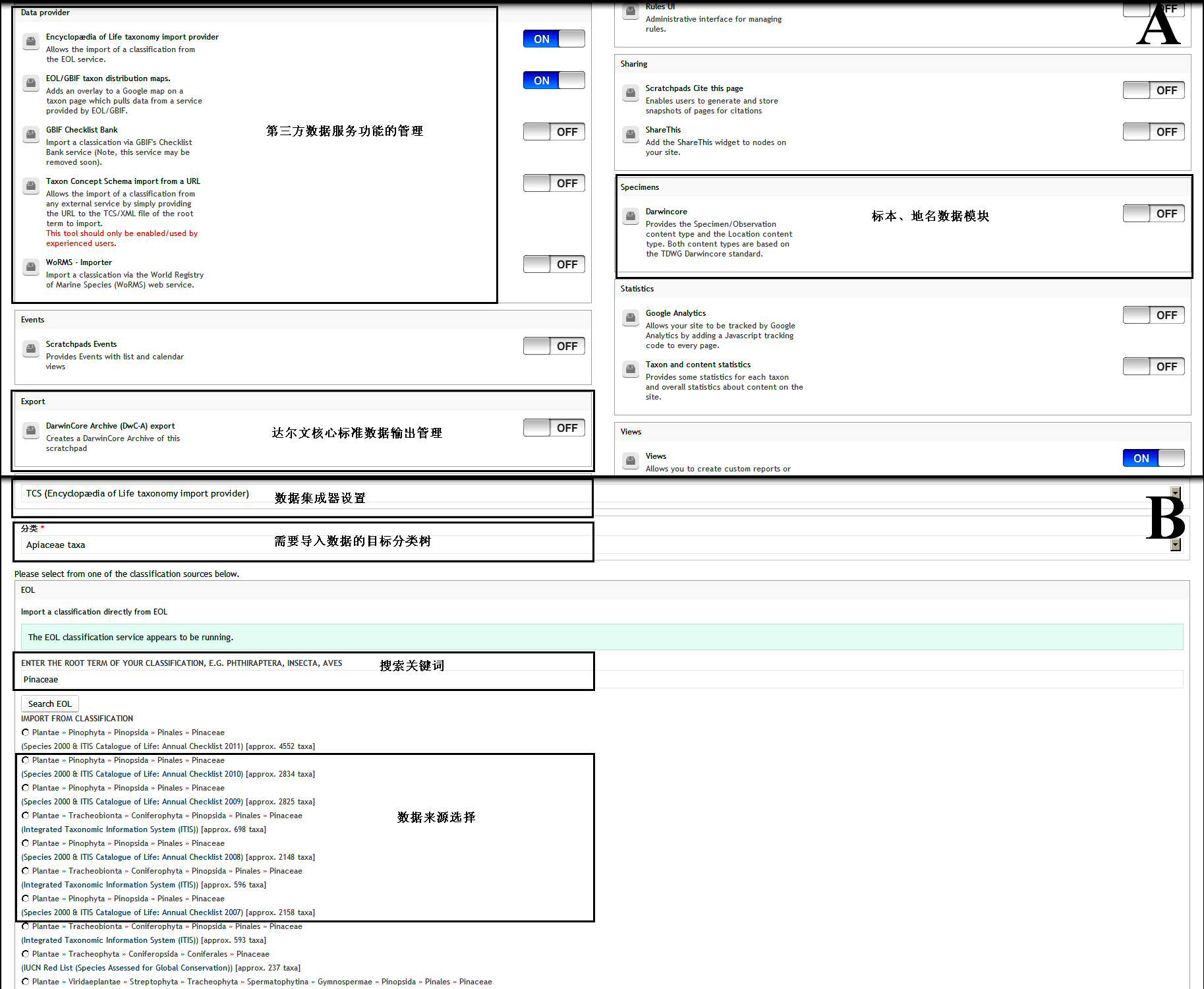
Fig. S1 Visualized interface of module management (A) and automatically taxonomic data import (B) in Scratchpads 2.0 http://www.biodiversity-science.net/fileup/PDF/w2014-012-1.pdf
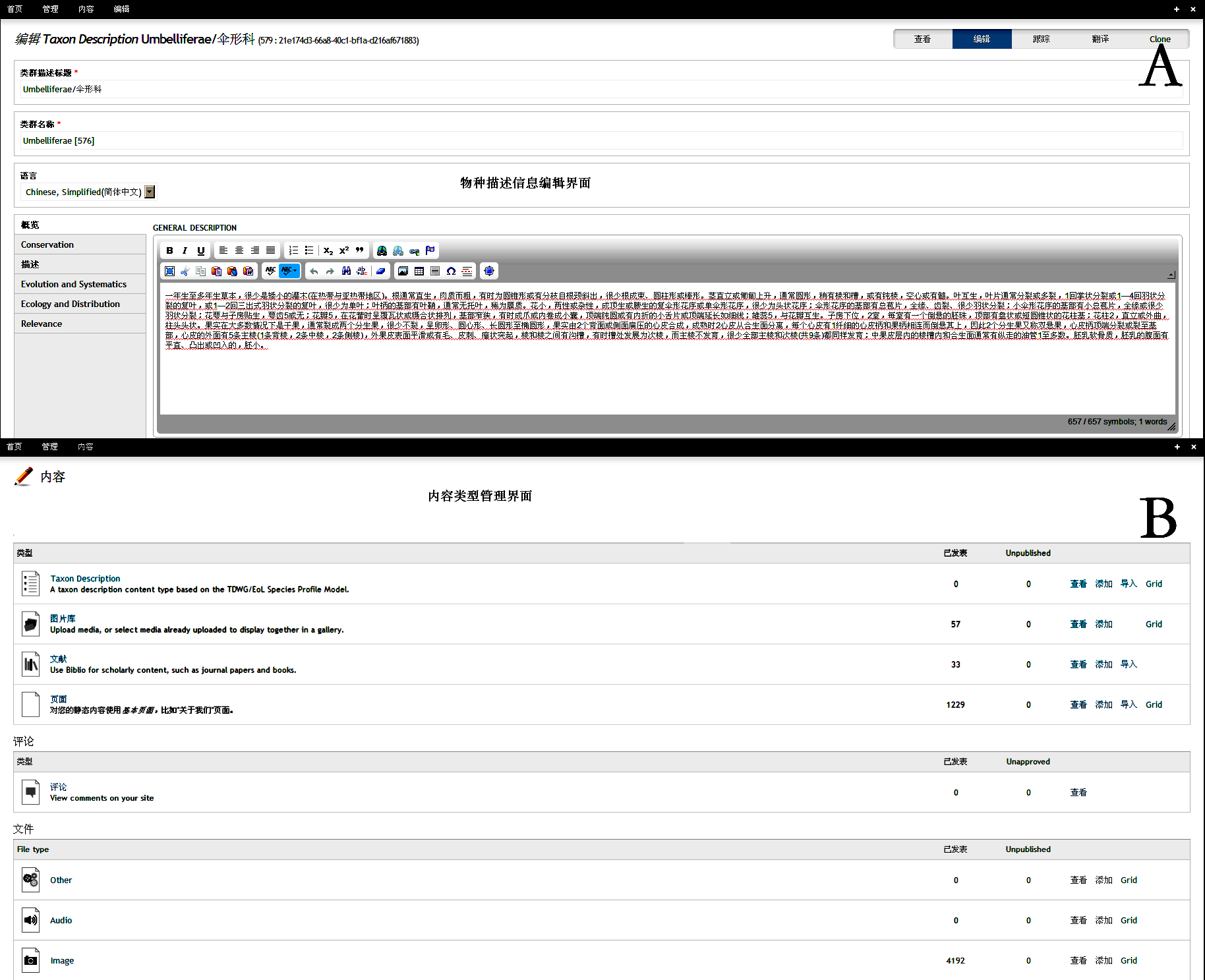
Fig. S3 Visulized interface of species description editor (A) and content management (B) in Scratchpads 2.0 http://www.biodiversity-science.net/fileup/PDF/w2014-012-3.pdf
| [1] | Agosti D (2003) Encyclopedia of life: Should species description equal gene sequence?Trends in Ecology and Evolution, 18, 273. |
| [2] | Baker E, Rycroft S, Smith VS (2014) Linking multiple biodiversity informatics platforms with Darwin Core Archives.Biodiversity Data Journal, 2, e1039. doi: 10.3897/BDJ.2. e1039. |
| [3] | Bebber DP, Carine MA, Wood JRI, Wortley AH, Harris DJ, Prance GT, Davidse G, Paige J, Pennington TD, Robson NKB, Scotland RW (2010) Herbaria are a major frontier for species discovery. Proceedings of the National Academy of Sciences,USA, 107, 22169-22171. |
| [4] | Benson DA, Karsch-Mizra CI, Lipman DJ, Ostell J, Sayers EW (2011) GenBank. Nucleic Acids Research, January; 39 (Database issue): D32-D37. Published online 2010 November 10. doi: 10.1093/nar/gkq1079. |
| [5] | Blagoderov V, Brake I, Georgiev T, Penev L, Roberts D, Rycroft S, Scott B, Agosti D, Catapano T, Smith VS (2010) Streamlining taxonomic publication: a working example with Scratchpads and ZooKeys.ZooKeys, 50, 17-28. |
| [6] | Bowker GC (2000) Biodiversity datadiversity.Social Studies of Science, 30, 643-683. |
| [7] | Brach AR, Boufford DE (2011) Why are we still producing paper floras?Annals of the Missouri Botanical Garden, 98, 297-300. |
| [8] | Brake I, Duin D, Van de Velde I, Smith V, Rycroft S (2011) Who learns from whom? Supporting users and developers of a major biodiversity e-infrastructure.ZooKeys, 150, 177-192. |
| [9] | Chavan V, Ingwersen P (2009) Towards a data publishing framework for primary biodiversity data: challenges and potentials for the biodiversity informatics community.BMC Bioinformatics, 10, S2. |
| [10] | Chavan V, Penev L (2011) The data paper: a mechanism to incentivize data publishing in biodiversity science.BMC Bioinformatics, 12, S2. |
| [11] | Clark BR, Godfray HCJ, Kitching IJ, Mayo SJ, Scoble MJ (2009) Taxonomy as an e-Science. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences, 367, 953-966. |
| [12] | Costello MJ (2009) Motivating online publication of data.BioScience, 59, 418-427. |
| [13] | Costello MJ, May RM, Stork NE (2013a) Can we name Earth's species before they go extinct?Science, 339, 413-416. |
| [14] | Costello MJ, Michener WK, Gahegan M, Zhang Z-Q, Bourne PE (2013b) Biodiversity data should be published, cited, and peer reviewed.Trends in Ecology and Evolution, 28, 454-461. |
| [15] | Deans AR, Yoder MJ, Balhoff JP (2012) Time to change how we describe biodiversity.Trends in Ecology and Evolution, 27, 78-84. |
| [16] | Duin D, van den Besselaar P (2011) Studying the effects of virtual biodiversity research infrastructures.ZooKeys, 150, 193-210. |
| [17] | Erwin T, Stoev P, Georgiev T, Penev L (2011) ZooKeys 150: Three and a half years of innovative publishing and growth.ZooKeys, 150, 5-14. |
| [18] | Fontaine B, van Achterberg K, Alonso-Zarazaga MA, Araujo R, Asche M, Aspöck H, Aspöck U, Audisio P, Aukema B, Bailly N, Balsamo M, Bank RA, Belfiore C, Bogdanowicz W, Boxshall G, Burckhardt D, Chylarecki P, Deharveng L, Dubois A, Enghoff H, Fochetti R, Fontaine C, Gargominy O, Lopez MSG, Goujet D, Harvey MS, Heller K-G, van Helsdingen P, Hoch H, De Jong Y, Karsholt O, Los W, Magowski W, Massard JA, McInnes SJ, Mendes LF, Mey E, Michelsen V, Minelli A, Nafria JMN, van Nieukerken EJ, Pape T, De Prins W, Ramos M, Ricci C, Roselaar C, Rota E, Segers H, Timm T, van Tol J, Bouchet P (2012) New species in the Old World: Europe as a frontier in diodiversity exploration, a test bed for 21st century taxonomy.PLoS ONE, 7, e36881. |
| [19] | Frodin DG (2001) Guide to Standard Floras of the World. Cambridge University Press, Cambridge. |
| [20] | Godfray HCJ (2002) Challenges for taxonomy: the discipline will have to reinvent itself if it is to survive and flourish.Nature, 417, 17-19. |
| [21] | Godfray HCJ, Clark BR, Kitching IJ, Mayo SJ, Scoble MJ (2007) The web and the structure of taxonomy.Systematic Biology, 56, 943-955. |
| [22] | Hebert PDN, Cywinska A, Ball SL, de Waard JR (2003) Biological identifications through DNA barcodes.Proceedings of the Royal Society of London, Series B: Biological Sciences, 270, 313-321. |
| [23] | Johnson NF (2012) A collaborative, integrated and electronic future for taxonomy.Invertebrate Systematics, 25, 471-475. |
| [24] | Knapp S (2008) Taxonomy as a team sport. In: The New Taxonomy (ed. Wheeler Q), pp. 33-53. CRC Press Taylor & Francis Group, Boca Raton, London, New York. |
| [25] | Kress WJ (2004) Paper floras: how long will they last? a review of flowering plants of the Neotropics.American Journal of Botany, 91, 2124-2127. |
| [26] | Leptin M (2012) Open access—pass the buck.Science, 335, 1279. |
| [27] | Li GJ (李国杰), Chen XQ (程学旗) (2012) Big Data: a significant strategic area in future science, technology, and economy development—research status and thinking of Big Data. Bulletin of the Chinese Academy of Sciences(中国科学院院刊), 27, 11. |
| [28] | Lynch C (2008) Big data: How do your data grow?Nature, 455, 28-29. |
| [29] | Maddison DR, Guralnick R, Hill A, Reysenbach A-L, McDade LA (2012) Ramping up biodiversity discovery via online quantum contributions.Trends in Ecology and Evolution, 27, 72-77. |
| [30] | Marhold K, Stuessy T, Agababian M, Agosti D, Alford MH, Crespo A, Crisci JV, Dorr LJ, Ferencová Z, Frodin D, Geltman DV, Kilian N, Linder HP, Lohmann LG, Oberprieler C, Penev L, Smith GF, Thomas W, Tulig M, Turland N, Zhang XC (2013) The future of botanical monography: Report from an international workshop, 12-16 March 2012, Smolenice, Slovak Republic.Taxon, 62, 4-20. |
| [31] | Mayo SJ (2008) Alpha e-taxonomy: responses from the systematics community to the biodiversity crisis.Kew Bulletin, 63, 1-16. |
| [32] | Mitch W (2008) Big data: Wikiomics.Nature, 455, 22-25. |
| [33] | Moore W (2011) Biology needs cyberinfrastructure to facilitate specimen-level data acquisition for insects and other hyperdiverse groups.ZooKeys, 147, 479-486. |
| [34] | Parr CS, Guralnick R, Cellinese N, Page RDM (2012) Evolutionary informatics: unifying knowledge about the diversity of life.Trends in Ecology and Evolution, 27, 94-103. |
| [35] | Patterson DJ, Cooper J, Kirk PM, Pyle RL, Remsen DP (2010) Names are key to the big new biology.Trends in Ecology and Evolution, 25, 686-691. |
| [36] | Penev L, Kress WJ, Knapp S, Li DZ, Renner S (2010a) Fast, linked, and open―the future of taxonomic publishing for plants: launching the journal PhytoKeys.PhytoKeys, 1, 1-14. |
| [37] | Penev L, Roberts D, Smith VS, Agosti D, Erwin T (2010b) Taxonomy shifts up a gear: new publishing tools to accelerate biodiversity research. ZooKeys, 50, i-iv. |
| [38] | Penev L, Sharkey M, Erwin T, van Noort S, Buffington M, Seltmann K, Johnson N, Taylor M, Thompson F, Dallwitz M (2009) Data publication and dissemination of interactive keys under the open access model.ZooKeys, 21, 1-17. |
| [39] | Platnick NI (2013) The information content of taxon names: a reply to de Queiroz and Donoghue.Systematic Biology, 62, 175-176. |
| [40] | Scotland RW, Wood JRI (2012) Accelerating the pace of taxonomy.Trends in Ecology and Evolution, 27, 415-416. |
| [41] | Sheng YY (盛杨燕), Zhou T (周涛) (2013) Big Data Time:A Revolution that Will Transform How We Live, Work, and Think (大数据时代: 生活、工作与思维的大变革). Zhejiang People's Publishing House. (in Chinese) |
| [42] | Smith V (2009) Data publication: towards a database of everything.BMC Research Notes, 2, 113. |
| [43] | Smith V, Penev L (2011) Collaborative electronic infrastructures to accelerate taxonomic research.ZooKeys, 150, 1-3. |
| [44] | Smith V, Rycroft S, Brake I, Scott B, Baker E, Livermore L, Blagoderov V, Roberts D (2011) Scratchpads 2.0: a Virtual Research Environment supporting scholarly collaboration, communication and data publication in biodiversity science.ZooKeys, 150, 53-70. |
| [45] | Smith V, Rycroft S, Harman K, Scott B, Roberts D (2009) Scratchpads: a data-publishing framework to build, share and manage information on the diversity of life.BMC Bioinformatics, 10, S6. |
| [46] | Stoltzfus A, O’Meara B, Whitacre J, Mounce R, Gillespie E, Kumar S, Rosauer D, Vos R (2012) Sharing and re-use of phylogenetic trees (and associated data) to facilitate synthesis.BMC Research Notes, 5, 1-15. |
| [47] | Stuessy TF (2009) Paradigms in biological classification (1707-2007): Has anything really changed?Taxon, 58, 68-76. |
| [48] | Thessen A, Patterson D (2011) Data issues in the life sciences.ZooKeys, 150, 15-51. |
| [49] | Thomas C (2009) Biodiversity databases spread, prompting unification call.Science, 324, 1632-1633. |
| [50] | Thomas WW, Forzza RC, Michelangeli FA, Giulietti AM, Leitman PM (2011) Large-scale monographs and floras: the sum of local floristic research.Plant Ecology and Diversity, 5, 217-223. |
| [51] | Vision T (2010) Open data and the social contract of scientific publishing.BioScience, 60, 330-331. |
| [52] | Whitlock MC (2011) Data archiving in ecology and evolution: best practices.Trends in Ecology and Evolution, 26, 61-65. |
| [53] | Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M, Giovanni R, Robertson T, Vieglais D (2012) Darwin Core: an evolving community-developed biodiversity data standard.PLoS ONE, 7, e29715. |
| [54] | Winker K, Withrow JJ (2013) Natural history: small collections make a big impact.Nature, 493, 480. |
| [55] | Yesson C (2007) How Global Is the Global Biodiversity Information Facility?PLoS ONE, 2, e1124. |
| [1] | Chenchen Ding, Dongni Liang, Wenpei Xin, Chunwang Li, Eric I. Ameca, Zhigang Jiang. A dataset on the morphological, life-history and ecological traits of the mammals in China [J]. Biodiv Sci, 2022, 30(2): 21520-. |
| [2] | Tongyi Liu, Jing Chen, Liyun Jiang, Gexia Qiao. Annual report of new taxa for Chinese Hemiptera and 28 other orders of Insecta in 2020 [J]. Biodiv Sci, 2021, 29(8): 1050-1057. |
| [3] | Fenglin Zhang,Xin Wang,Jian Zhang. Biodiversity information resources. II. Environmental data [J]. Biodiv Sci, 2018, 26(1): 53-65. |
| [4] | Jian Zhang. Biodiversity science and macroecology in the era of big data [J]. Biodiv Sci, 2017, 25(4): 355-363. |
| [5] | Xin Wang, Fenglin Zhang, Jian Zhang. Biodiversity information resources. I. Species distribution, catalogue, phylogeny, and life history traits [J]. Biodiv Sci, 2017, 25(11): 1223-1238. |
| [6] | Kwangtsao Shao, Han Lee, Yungchang Lin, Kunchi Lai. A review of marine biodiversity information resources [J]. Biodiv Sci, 2014, 22(3): 253-263. |
| [7] | Kunchi Lai, Youhua Cheng, Yuehchih Chen, Yousheng Li, Kwangtsao Shao. Applying cluster analysis and Google Maps in the study of large-scale species occurrence data [J]. Biodiv Sci, 2012, 20(1): 76-85. |
| [8] | Kwangtsao Shao, Kunchi Lai, Yungchang Lin, Chihjen Ko, Han Lee, Lingya Hung, Yuehchih Chen, Leesea Chen. Experience and strategy of biodiversity data integration in Taiwan [J]. Biodiv Sci, 2010, 18(5): 444-453. |
| [9] | Zheping Xu, Jinzhong Cui, Haining Qin, Keping Ma. On the architecture of biodiversity e-Science infrastructure in China [J]. Biodiv Sci, 2010, 18(5): 480-488. |
| [10] | ZHONG Yang, ZHANG Liang, REN Wen-Wei, CHEN Jia-Kuan. Biodiversity Informatics: a new direction of bioinformatics and biodiversity science and related key techniques [J]. Biodiv Sci, 2000, 08(4): 397-404. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn