滨海湿地位于淡水与海水交汇的过渡地带, 具有维持生物多样性等多种生态服务功能(李伟等, 2014)。水文连通作为湿地生态过程的主要驱动因素之一, 是指在水文循环要素内部或要素之间以水为媒介的物质、能量和生物的传输(Freeman et al, 2007)。水文连通在塑造地形结构, 物质运输, 调控植被组成和有机质积累的时空格局中发挥重要作用(Yuan et al, 2015), 通过直接作用于无机环境中的物质循环和能量流动进而影响初级生产力, 然后基于生物级联效应对食物链中其他营养级造成影响, 改变鱼类及其他生物类群的食源结构(Jardine et al, 2012; 崔保山等, 2016)。水文连通还能影响鱼类迁移及其生殖细胞在水中的传输进而影响其分布特征与多样性(Schofield et al, 2018)。黄河三角洲位于暖温带海陆交接地带, 是该地区保存最为完整、面积最为广阔的新生湿地生态系统。近年来, 三角洲湿地生态系统结构与功能均衡状态被打破, 面临退化危机(孙志高等, 2011)。气候变化以及人类活动的影响使滨海湿地水文连通格局发生了改变, 一定程度上影响了潮沟发育和湿地地下水系的流动, 导致水文连通性受阻且变差(赵心怡等, 2019)。目前, 许多学者围绕黄河三角洲水文连通的生态效应展开相关研究, 但大多都基于传统的多样性调查方法。
传统的鱼类多样性调查采样强度大、易受物理条件限制, 还要求调查人员拥有专业的分类鉴定知识, 所以往往会低估甚至误判研究区域内的生物多样性。因此, 亟需一种精准可靠、灵敏高效、快速且成本低的多样性调查手段结合传统形态学鉴定, 以实现对鱼类群落波动变化的准确监测, 从而为湿地生态系统的保护与管理提供更全面的支持。环境DNA宏条形码技术(eDNA metabarcoding)就是在这样的背景下发展起来的新型检测手段。环境DNA宏条形码技术是指对环境样本中组成复杂、来源广泛的游离DNA进行高通量测序, 然后比对参考数据库对测序序列进行注释分类, 是进行多物种检测、评估物种丰富度的有效方法(Creer et al, 2016)。近年来, 该技术已被广泛应用于河流(Sales et al, 2021; 沈梅等, 2022)、湖泊(Sard et al, 2019)、河口(Kume et al, 2021)及海洋(Murakami et al, 2019)等各类水生生态系统鱼类多样性监测中。鱼类常用的遗传标记有12S rRNA基因(Miya et al, 2015; Valentini et al, 2016)、16S rRNA基因(Evans et al, 2016; Shaw et al, 2016)、COI基因(Balasingham et al, 2018)、Cytb基因(Miya & Nishida, 2000; Thomsen et al, 2012b)等。目前, 基于环境DNA技术对黄河三角洲地区开展鱼类多样性监测以及探讨环境因子对其影响机制尚未见报道。
本研究选择黄河三角洲具有明显水文连通梯度的典型潮沟系统, 采用环境DNA宏条形码技术进行河口湿地鱼类多样性研究, 旨在明确河口典型潮沟系统鱼类多样性, 同时分析不同水文连通梯度潮沟生态环境特征及其与环境DNA表征的鱼类多样性之间的关系, 明确鱼类eDNA多样性对潮沟水文连通特征的响应, 为黄河口湿地鱼类生物多样性监测以及水生生态保护提供新的技术方法。
1 材料与方法
1.1 研究区域概况与采样站位设置
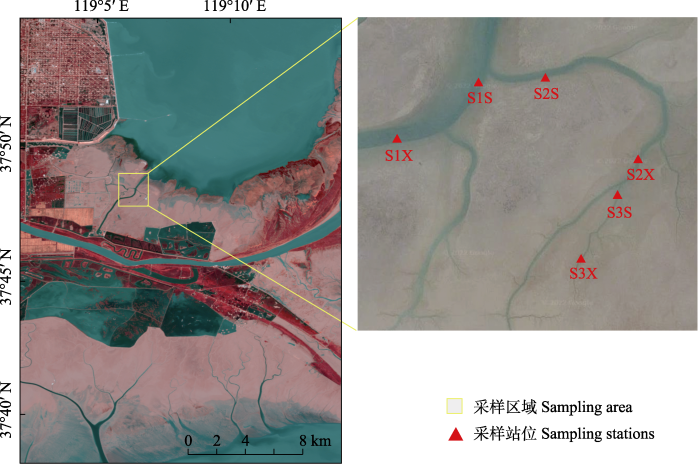
研究区域位于山东黄河三角洲国家级自然保护区内, 该区域地处黄河入海口, 面朝渤海, 南邻莱州湾, 是中国暖温带保存最完整、最广阔、最年轻的湿地生态系统, 属季风气候区, 区域内滩涂湿地遍布, 潮沟水系发达。在保护区北部区域选择一条完整的、具有明显水文连通梯度的潮沟设置采样站位, 地理坐标为37°28′-37°29′ N, 119°03′-119°03′ E。根据潮沟的主次支流进行分级, 与海直接相连且断面面积最大的为一级潮沟, 其支流为二级潮沟, 次支流末端不再分支且流量最小的为三级潮沟(李冬雪等, 2020), 分别在每级潮沟的上下游设置样点, 共布设6个样点(图1)。使用TCM-4浅水倾斜流速仪海流计测量各级别潮沟流速, 一级潮沟至三级潮沟流速分别为21.32 cm/s、14.70 cm/s、3.75 cm/s, 潮沟流速与断面面积直接相关, 结合现场勘察也发现一级潮沟至三级潮沟宽度、水深依次递减。
图1
图1
黄河口自然湿地采样区域。S1S: 一级潮沟上游; S1X: 一级潮沟下游; S2S: 二级潮沟上游; S2X: 二级潮沟下游; S3S: 三级潮沟上游; S3X: 三级潮沟下游。
Fig. 1
Sampling area of natural wetland in the Yellow River Delta. S1S, Upstream of the first-class tidal creek; S1X, Downstream of the first-class tidal creek; S2S, Upstream of the second-class tidal creek; S2X, Downstream of the second-class tidal creek; S3S, Upstream of the third-class tidal creek; S3X, Downstream of the third-class tidal creek.
1.2 水样采集与环境因子测定
于2022年9月27日进行水样采集, 各样点采集潮沟内表层水样1 L, 沿岸每隔5 m用1 L瓶盛取1/3, 最后装满。采集后暂时保存于-20℃车载冰箱中, 24 h内带回实验室, 用三联真空抽滤装置内装0.45 μm滤膜抽滤水样。抽滤每份样品前使用75%医用酒精清洁滤瓶并用蒸馏水润洗以防止交叉污染, 以过滤等量蒸馏水作为阴性对照, 所得滤膜在提取环境DNA前于-80 ℃保存。
采用YSI (Professional Plus (6050000)/ProComm (605604), 美国)现场同步记录水体温度(T)、盐度(SAL)、溶解氧(DO)、酸碱度(pH值)等水质参数。另取部分水样带回实验室经0.45 μm滤膜过滤后使用连续流动分析仪(SEAL, 德国)测定水体磷酸盐(PO43--P)、硝酸盐(NO3--N)、亚硝酸盐(NO2--N)、铵盐(NH4+-N)、硅酸盐(SiO32--Si)含量以及总氮(TN)、总磷(TP)含量, 测定按照《海洋调查规范》(GB/T12763.4-2007)进行。
1.3 环境DNA提取、PCR扩增与高通量测序
使用Fast DNATM SPIN Kit for Soil (MP Biomedicals, 美国)提取滤膜DNA。使用Nanodrop One (Thermo Scientific, 美国)测定DNA溶液浓度及纯度(A260/A280: 1.6-2.0; 浓度 ≥ 10 ng/μL), 并进行1%浓度的琼脂糖凝胶电泳检测。
选择鱼类线粒体12S rRNA基因通用引物(Miya et al, 2015): MiFish-E-F: 5'-GTTGGTAAATCTCGT GCCAGC-3'和MiFish-E-R: 5'-CATAGTGGGGTATC TAATCCTAGTTTG-3', 对环境DNA样品进行PCR扩增。引物由生工生物工程(上海)股份有限公司合成, 基于引物Tm值设置温度梯度PCR确定合适的退火温度。20 μL PCR反应体系: 5 × FastPfu Buffer 4 μL、2.5 mM dNTPs 2 μL、正反引物(5 μM)各0.8 μL、FastPfu Polymerase 0.4 μL、模板DNA 10 ng、补ddH2O至20 μL。PCR反应参数: 95℃预变性5 min; 95℃变性30 s, 55℃退火30 s, 72℃延伸45 s, 进行27个循环; 最终于72℃下延伸10 min。所用PCR仪为ABI GeneAmp® 9700型。每个样品3个技术重复, 同一样品的PCR产物混合后用2%琼脂糖凝胶电泳检测, 使用AxyPrepDNA凝胶回收试剂盒(AXYGEN, 美国)切胶回收PCR产物, 送至上海凌恩生物科技有限公司于Illumina NovaSeq 6000测序平台(Illumina, 美国)进行高通量测序。
1.4 数据处理及分析
使用软件SPSS 26对各级别潮沟环境理化指标进行显著性检验, 若数据满足正态分布且符合方差齐性, 显著性水平以单因素方差分析(analysis of variance, ANOVA)检验为最终结果, 多重比较选择Bonferroni检验; 若数据满足正态分布但方差不齐, 以Welch检验为显著性水平最终结果, 多重比较选择Tamhane T2检验; 若数据不满足正态分布, 使用独立样本的非参数检验, 然后通过Bonferroni校正法调整显著性值。
根据barcode序列和PCR扩增引物序列从下机数据中拆分出各样本数据, 截去barcode和引物序列后基于序列间的overlap关系, 对成对reads进行序列拼接, 再经质控、去除嵌合体后得到有效序列。对有效序列提取非重复序列, 去除单序列, 然后按照97%相似性对非重复序列进行OTU聚类, 同时筛选OTUs中出现频数最高的序列作为OTUs的代表序列, 将所有优化序列map至OTU代表序列, 生成OTU表格。使用R包的dplyr函数基于样本丰度矩阵, 将各样本中reads小于100的OTUs剔除; 将所得OTUs的fasta序列文件导入NCBI进行blastn比对(与nt库进行比对), 保留identity > 90%、query coverage > 85%的OTUs (Zhan et al, 2013), 再基于identity、query coverage以及e-value 3项指标筛选出每个OTU比对结果中最相似的结果对其进行注释。利用世界海洋物种登记库(WoRMS) (
2 结果
2.1 各级别潮沟生态环境特征的空间差异
测定的13种环境因子指标如表1所示。通过两两比较发现, 在不同级别的潮沟组合之间, 各环境因子均至少在一组级别间存在显著差异(P < 0.05)。除总磷外, 其余各环境指标在一级潮沟与三级潮沟之间均存在显著差异(P < 0.05), 氧化还原电位、磷酸盐、电导率、盐度以及pH值存在极显著差异(P < 0.01)。一级潮沟的硝酸盐、磷酸盐、pH值和溶解氧的含量显著高于三级潮沟, 但三级潮沟的总氮、亚硝酸盐、硅酸盐、电导率、盐度均显著高于一级潮沟; 另外, 二级潮沟的总磷浓度显著低于其余两级潮沟。部分环境因子由一级潮沟至三级潮沟呈现出递增或递减的趋势。例如, 从一级潮沟到三级潮沟, 总氮、亚硝酸盐、硅酸盐、电导率以及盐度含量逐级升高, 表现为S1 < S2 < S3 (P > 0.05); 磷酸盐、铵盐、硝酸盐、溶解氧、pH值以及氧化还原电位逐级降低, 表现为S1 > S2 > S3 (P > 0.05)。
表1 黄河口潮沟环境理化指标。同行不同小写字母表示差异显著(P < 0.05)。加粗表示最大值。
Table 1
| 变量 Variable | 潮沟级别 Tidal creek level | ||
|---|---|---|---|
| 一级潮沟 The first-class tidal creek (S1) | 二级潮沟 The second-class tidal creek (S2) | 三级潮沟 The third-class tidal creek (S3) | |
| 总氮 TN (μg/L) | 351.893 ± 4.857b | 361.896 ± 11.934ab | 389.191 ± 19.168a |
| 总磷 TP (μg/L) | 15.116 ± 1.175a | 12.586 ± 0.580b | 14.975 ± 0.715a |
| 磷酸盐 PO43--P (mg/L) | 0.033 ± 0.029a | 0.008 ± 0.002ab | 0.004 ± 0.001b |
| 铵盐 NH4+-N (mg/L) | 0.039 ± 0.012a | 0.036 ± 0.004a | 0.170 ± 0.121b |
| 亚硝酸盐 NO2--N (mg/L) | 0.019 ± 0.003b | 0.024 ± 0.004ab | 0.027 ± 0.004a |
| 硅酸盐 SiO32--Si (mg/L) | 0.675 ± 0.052b | 0.690 ± 0.107ab | 0.964 ± 0.222a |
| 硝酸盐 NO3--N (mg/L) | 0.265 ± 0.041a | 0.238 ± 0.018a | 0.193 ± 0.017b |
| 温度 T (℃) | 21.800 ± 1.595b | 21.450 ± 0.321b | 26.233 ± 2.850a |
| 溶解氧 DO (mg/L) | 7.998 ± 0.804a | 6.692 ± 0.315b | 6.067 ± 1.143b |
| 电导率 SPC (ms/cm) | 26.412 ± 0.245b | 32.538 ± 1.752ab | 36.300 ± 1.715a |
| 盐度 SAL | 16.185 ± 0.170b | 20.362 ± 1.202ab | 22.903 ± 1.121a |
| 酸碱度 pH | 8.408 ± 0.012a | 8.385 ± 0.021ab | 8.248 ± 0.026b |
| 氧化还原电位 Eh | -71.950 ± 1.104a | -70.583 ± 1.087a | -64.033 ± 1.219b |
2.2 高通量测序基础信息
2022年9月黄河口潮沟水样环境DNA高通量测序结果见表2。共送测18个样品, 2%琼脂糖凝胶电泳检测纯化后PCR产物, 样品目的条带弱, 为更好满足后续建库上机要求, 将同站位3个平行样合为1个, 实际测序6个样品。测序原始下机产生的双端序列, 经质控过滤、拼接并去除嵌合体后, 最终得到可用于后续分析的有效序列, 共3,122,115条, 碱基数为529,223,294, 序列均长169.51 bp, 读长位于151-200 bp之间的序列占比为99.54%。6个样品聚类得到3,314个OTUs。
表2 各样品优化序列统计
Table 2
| 样品名称 Sample name | 序列条数 Sequences | 碱基数 Bases (bp) | 平均读长 Average length (bp) |
|---|---|---|---|
| S1S | 500,311 | 85,254,665 | 170.40 |
| S1X | 500,256 | 84,418,677 | 168.75 |
| S2S | 471,672 | 80,731,476 | 171.16 |
| S2X | 479,424 | 81,634,922 | 170.28 |
| S3S | 556,415 | 93,868,748 | 168.70 |
| S3X | 614,037 | 103,314,806 | 168.26 |
2.3 物种组成与空间分布
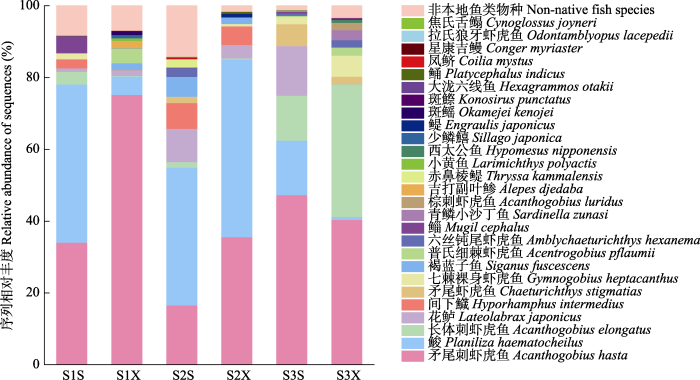
根据样品序列丰度, 将不符合要求的OTUs除去, 再筛除掉与NCBI核酸数据库比对结果中未满足条件的OTUs, 最终保留了489个OTUs, 能注释到3个门、6个纲、25个目、37个科、61个属、68个种。其中鱼类有55种, 隶属于2纲12目27科48属, 检测到的鱼类中有27种属于黄河口、莱州湾水域本地物种, 隶属于2纲9目16科25属(表3)。本地鱼类在目水平上物种数排名前三的分别为鲈形目13种、鲱形目5种、鲻形目与鲉形目各2种; 在科水平上物种数排名前三的分别为虾虎鱼科8种、鳀科3种、鲱科与鲻科各2种; 序列丰度排名前三的物种分别为矛尾刺虾虎鱼(Acanthogobius hasta)、鮻(Planiliza haemato- cheilus)以及长体刺虾虎鱼(Acanthogobius elongatus) (图2)。不同级别潮沟在物种组成与序列丰度上呈现出一定程度的空间分布差异(图3), 其中矛尾刺虾虎鱼在各样点均被检测到, 且在一级潮沟下游有着最高的序列丰度; 长体刺虾虎鱼序列丰度在三级潮沟最高; 非本地鱼类物种序列仅在主潮沟及其邻近样点有分布。
表3 基于环境DNA宏条形码在黄河口潮沟中检测到的鱼类物种。“本地鱼类”一词指代的是那些在黄河口、莱州湾海域历史渔业资源调查中实际捕获到或者曾经捕获到的海洋鱼类。
Table 3
| 目 Order | 科 Family | 种 Species | |
|---|---|---|---|
| 本地鱼类物种 | 鲽形目 Pleuronectiformes | 舌鳎科 Cynoglossidae | 短吻红舌鳎 Cynoglossus joyneri |
| Native fish species | 鲱形目 Clupeiformes | 鲱科 Dorosomatidae | 青鳞小沙丁鱼 Sardinella zunasi |
| 斑鰶 Konosirus punctatus | |||
| 鳀科 Engraulidae | 赤鼻棱鳀 Thryssa kammalensis | ||
| 鳀 Engraulis japonicus | |||
| 凤鲚 Coilia mystus | |||
| 颌针鱼目 Beloniforme | 鱵科 Hemiramphidae | 间下鱵 Hyporhamphus intermedius | |
| 胡瓜鱼目 Osmeriformes | 胡瓜鱼科 Osmeridae | 西太公鱼 Hypomesus nipponensis | |
| 鲈形目 Perciformes | 花鲈科 Lateolabracidae | 花鲈 Lateolabrax japonicus | |
| 蓝子鱼科 Siganidae | 褐蓝子鱼 Siganus fuscescens | ||
| 鲹科 Carangidae | 吉打副叶鲹 Alepes djedaba | ||
| 石首鱼科 Sciaenidae | 小黄鱼 Larimichthys polyactis | ||
| 鱚科 Sillaginidae | 少鳞鱚 Sillago japonica | ||
| 虾虎鱼科 Gobiidae | 矛尾刺虾虎鱼 Acanthogobius hasta | ||
| 长体刺虾虎鱼 Acanthogobius elongatus | |||
| 矛尾虾虎鱼 Chaeturichthys stigmatias | |||
| 七棘裸身虾虎鱼 Gymnogobius heptacanthus | |||
| 普氏细棘虾虎鱼 Acentrogobius pflaumii | |||
| 六丝钝尾虾虎鱼 Amblychaeturichthys hexanema | |||
| 棕刺虾虎鱼 Acanthogobius luridus | |||
| 拉氏狼牙虾虎鱼 Odontamblyopus lacepedii | |||
| 鳗鲡目 Anguilliformes | 康吉鳗科 Congridae | 星康吉鳗 Conger myriaster | |
| 鳐形目 Rajiformes | 鳐科 Rajidae | 斑鳐 Okamejei kenojei | |
| 鲉形目 Scorpaeniformes | 六线鱼科 Hexagrammidae | 大泷六线鱼 Hexagrammos otakii | |
| 鲬科 Platycephalidae | 鲬 Platycephalus indicus | ||
| 鲻形目 Mugiliformes | 鲻科 Mugilidae | 鮻 Planiliza haematocheilus | |
| 鲻 Mugil cephalus | |||
| 非本地鱼类物种 | 鲱形目 Clupeiformes | 鲱科 Clupeidae | 隆背小沙丁鱼 Sardinella gibbosa |
| Non-native fish species | 花点鲥 Hilsa kelee | ||
| 鳉形目 Cyprinodontiformes | 青鳉科 Oryziatidae | 青鳉 Oryzias latipes | |
| 鲤形目 Cypriniformes | 鲤科 Cyprinidae | 鲫 Carassius auratus | |
| 似鳊 Pseudobrama simoni | |||
| 尖棘鱲 Zacco acutipinnis | |||
| 鲤 Cyprinus carpio | |||
| 温州光唇鱼 Acrossocheilus wenchowensis | |||
| 三角鲂 Megalobrama terminalis | |||
| 长鳍马口鱼 Opsariichthys evolans | |||
| 刺鲃 Spinibarbus caldwelli | |||
| 贝氏䱗 Hemiculter bleekeri | |||
| 爬鳅科 Balitoridae | 梅花山拟腹吸鳅 Pseudogastromyzon meihuashanensis | ||
| 原缨口鳅 Vanmanenia stenosoma | |||
| 鲈形目 Perciformes | 金线鱼科 Nemipteridae | 日本金线鱼 Nemipterus thosaporni | |
| 丽鱼科 Cichlidae | 齐氏非鲫 Coptodon zillii | ||
| 非鲫 Oreochromis mossambicus | |||
| 加利略帚齿非鲫 Sarotherodon galilaeus | |||
| 尼罗非鲫 Oreochromis niloticus | |||
| 雀鲷科 Pomacentridae | 白带金翅雀鲷 Chrysiptera brownriggii | ||
| 塘鳢科 Eleotridae | 刺盖塘鳢 Eleotris acanthopomus | ||
| 虾虎鱼科 Gobiidae | 波氏吻虾虎鱼 Rhinogobius cliffordpopei | ||
| 雀斑吻虾虎鱼 Rhinogobius lentiginis | |||
| 名古屋吻虾虎鱼 Rhinogobius nagoyae | |||
| 武义吻虾虎鱼 Rhinogobius wuyiensis | |||
| 鲇形目 Siluriformes | 鲿科 Bagridae | 黄颡鱼 Tachysurus fulvidraco | |
| 鲇科 Siluridae | 怀头鲇 Silurus soldatovi | ||
| 甲鲇科 Loricariidae | 豹纹脂身鲇 Pterygoplichthys pardalis |
图2
图2
各采样站位鱼类的序列相对丰度。采样站位缩写全称见
Fig. 2
The relative abundance of fish sequences at each sampling station. The full names of the sampling stations' abbreviations are shown in
图3
图3
各采样站位基于序列丰度的鱼类空间分布热图。采样站位缩写全称见
Fig. 3
Heatmap of the spatial distribution of fish species at each sampling station based on the abundance of sequences. The full names of the sampling stations' abbreviations are shown in
2.4 各级别潮沟鱼类生物多样性空间差异
基于55种鱼类序列相对丰度矩阵计算得到各样点鱼类群落α多样性指数(表4)。各样品文库覆盖率(Good's coverage)较高, 均位于99%以上, 表明样品序列检出率高, 测序结果真实可靠。Chao 1指数和ACE指数总体上表现为S2 > S1 > S3 (P > 0.05)。Shannon指数呈现出“两头低, 中间高”的分布趋势, 即二级潮沟的Shannon指数较一级和三级潮沟高; Simpson指数与Shannon指数变化趋势一致, 即二级潮沟的多样性指数最高。
表4 基于序列相对丰度计算得到的鱼类群落α多样性指数
Table 4
| 采样站点 Sampling station | 观察物种数 Observed species | ACE指数 ACE index | Chao 1指数 Chao 1 index | Shannon指数 Shannon index | Simpson指数 Simpson index | 测序深度指数 Good's coverage |
|---|---|---|---|---|---|---|
| S1S | 325 | 328.28 | 328.11 | 3.95 | 0.84 | 0.999977 |
| S1X | 327 | 329.57 | 328.62 | 3.14 | 0.62 | 0.999980 |
| S2S | 324 | 335.00 | 350.25 | 4.44 | 0.87 | 0.999958 |
| S2X | 316 | 325.00 | 323.50 | 3.25 | 0.73 | 0.999958 |
| S3S | 325 | 337.51 | 335.69 | 3.60 | 0.78 | 0.999946 |
| S3X | 288 | 302.85 | 302.88 | 3.64 | 0.81 | 0.999935 |
2.5 鱼类群落组成与水环境因子的关系
基于序列丰度排名前15位的本地鱼类物种丰度矩阵进行去趋势对应分析(detrended correspondence analysis, DCA), 根据排序轴梯度长度(gradient length < 3), 选择RDA分析。对环境数据(pH除外)进行lg(x + 1)转换。通过蒙特卡洛检验分析环境数据显著性, 根据Forward Selection结果筛选出显著相关的环境因子进行后续排序分析。蒙特卡洛检验结果表明, 第一轴和所有轴均具有显著性(P < 0.05), 即RDA排序结果可信。
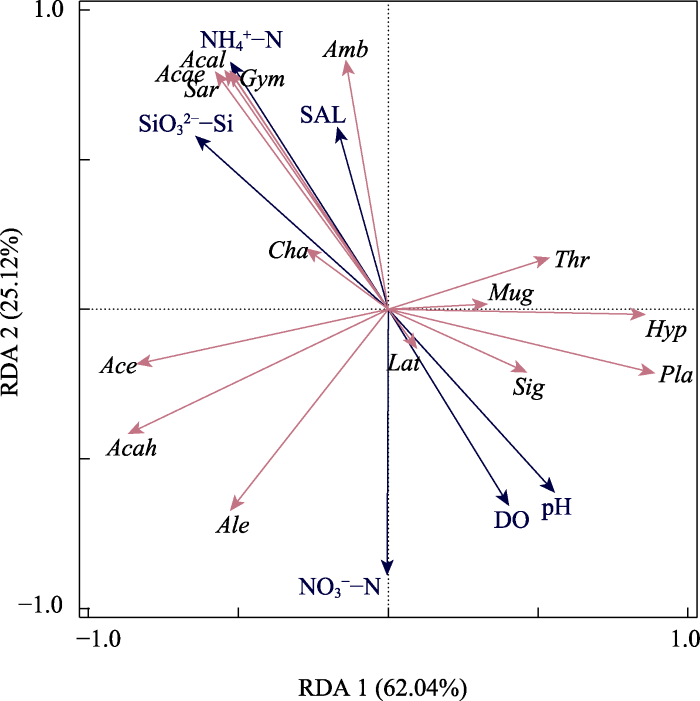
鱼类物种丰度矩阵与水环境因子的RDA分析结果如表5所示。第一轴和第二轴包含的物种与环境相关系数分别为0.969、0.992, 共解释了物种-环境关系总方差的91.7%。与鱼类群落结构显著相关(P < 0.05)的6种环境因子的解释率(conditional effects)分别为: 硅酸盐(SiO32--Si) 34.7%、硝酸盐(NO3--N) 21.3%、pH值15.0%、盐度(SAL) 13.8%、铵盐(NH4+-N) 6.2%、溶解氧(DO) 4.1%。RDA排序可以有效反映鱼类群落分布与环境因子之间的关系。
表5 黄河口潮沟鱼类物种序列丰度冗余分析(RDA)
Table 5
| 排序轴 Axes | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| 特征值 Eigenvalues | 0.62 | 0.251 | 0.046 | 0.028 |
| 相关系数 Species-environment correlations | 0.969 | 0.992 | 0.986 | 0.947 |
| 物种数据累积百分比方差 Cumulative percentage variance of species data (%) | 62 | 87.2 | 91.8 | 94.7 |
| 物种-环境关系累积百分比方差 Cumulative percentage variance of species-environment relation (%) | 65.3 | 91.7 | 96.6 | 99.6 |
| 总特征值 Sum of all eigenvalues | 1 | |||
| 总典范特征值 Sum of all canonical eigenvalues | 0.951 | |||
鱼类群落结构与环境因子之间关系的冗余分析(图4)显示, 虾虎鱼科7种虾虎鱼的序列丰度分布均与硅酸盐呈正相关, 与pH、DO呈负相关, 除矛尾刺虾虎鱼以外, 其余虾虎鱼的序列丰度分布与盐度呈正相关; 总体上, 虾虎鱼科鱼类序列丰度与其余绝大多数鱼类的序列丰度呈负相关。
图4
图4
冗余分析(RDA)探究鱼类群落结构与环境因子之间的关系。Acae: 长体刺虾虎鱼; Acah: 矛尾刺虾虎鱼; Acal: 棕刺虾虎鱼; Ace: 普氏细棘虾虎鱼; Ale: 吉打副叶鲹; Amb: 六丝钝尾虾虎鱼; Cha: 矛尾虾虎鱼; Gym: 七棘裸身虾虎鱼; Hyp: 间下鱵; Lat: 花鲈; Mug: 鲻; Pla: 鮻; Sar: 青鳞小沙丁鱼; Sig: 褐蓝子鱼; Thr: 赤鼻棱鳀。DO: 溶解氧; NH4+-N: 铵盐; NO3--N: 硝酸盐; pH: 酸碱度; SAL: 盐度; SiO32--Si: 硅酸盐。
Fig. 4
Redundancy analysis (RDA) exploring the relationship between fish community structure and measured environmental variables. Acae, Acanthogobius elongatus; Acah, Acanthogobius hasta; Acal, Acanthogobius luridus; Ace, Acentrogobius pflaumii; Ale, Alepes djedaba; Amb, Amblychaeturichthys hexanema; Cha, Chaeturichthys stigmatias; Gym, Gymnogobius heptacanthus; Hyp, Hyporhamphus intermedius; Lat, Lateolabrax japonicus; Mug, Mugil cephalus; Pla, Planiliza haematocheilus; Sar, Sardinella zunasi; Sig, Siganus fuscescens; Thr, Thryssa kammalensis. DO, dissolved oxygen; NH4+-N, Ammoniumate; NO3--N, Nitrate; pH, Potential of hydrogen; SAL, Salinity; SiO32--Si, Silicate.
3 讨论
3.1 基于环境DNA宏条形码技术检测的潮沟系统鱼类物种组成
本研究基于环境DNA宏条形码技术评估黄河口保护区北部一典型潮沟系统中的鱼类多样性, 共检测到鱼类55种, 其中有27种属于莱州湾、黄河口水域常见鱼类物种, 在相关研究区域文献资料以及以往的渔业资源调查中均有确切记录(王平等, 1999; 李晴, 2008①(①李晴 (2008) 中国黄渤海区虾虎鱼类. 硕士学位论文, 南昌大学, 南昌.); 于海婷, 2013②(②于海婷 (2013) 山东近海典型海湾河口渔业资源调查与生物群落结构分析. 硕士学位论文, 中国海洋大学, 山东青岛.); 刘静等, 2015)。孙雅妮(2022)③(③孙雅妮 (2022) 黄河口及邻近水域生境及渔业生物多样性变化. 硕士学位论文, 烟台大学, 山东烟台.)在黄河口及其邻近海域进行底拖网调查, 3个航次共捕获鱼类54种, 与本研究重叠物种15种, 相同采样月份重叠物种13种; 高涵(2021)④(④高涵 (2021) 黄河口及附近海域渔业生物多样性与生境研究. 硕士学位论文, 烟台大学, 山东烟台.)通过3次底拖网调查, 共捕获鱼类46种, 与本研究重叠物种13种, 相同采样月份重叠物种8种; 李凡等(2013)统计的莱州湾生物量前10位渔业生物中的所有鱼类以及孙鹏飞等(2014)统计的莱州湾及黄河口水域除绯䲗(Callionymus beniteguri)以外的秋季鱼类重要种类在本研究中均被检测到。
一些传统底拖网调查中常见的鱼类, 如绯䲗、蓝点马鲛(Scomberomorus niphonius)等, 在本研究中未被检测到, 推测与采样季节和区域设置有关。绯䲗为冬、春季节优势种且多集中分布于莱州湾中部, 蓝点马鲛在产卵生殖季节(4-5月)会洄游至莱州湾河口及内湾, 但该鱼类为夏季优势种, 并且分布分散, 倾向于在较深水域产卵(李凡等, 2018; 瞿俊跃等, 2021)。而本研究采样区域为滩涂湿地潮沟水系, 区域较为局部, 且采样季节为秋季, 非鱼类产卵热季。本文中检测到的本地鱼类物种, 如虾虎鱼科物种, 多为小型底层鱼类, 喜栖息于近岸浅水及河口咸、淡水滩涂淤泥底质水域; 部分广盐或广温性鱼类, 如鲻(Mugil cephalus)、鳀(Engraulis japonicus), 也喜欢栖息于咸淡水交汇处; 还有部分洄游性鱼类, 如青鳞小沙丁鱼(Sardinella zunasi)、斑鰶(Konosirus punctatus)等, 会因为索饵等原因洄游至河口, 且在夏、秋季广泛分布于莱州湾(李凡等, 2018)。
本研究检测到的28种非本地鱼类, 物种组成主要由鲤科各属、丽鱼科各非鲫属、虾虎鱼科吻虾虎鱼属(Rhinogobius)等类群组成。检测到这些物种的原因是多方面的。首先, 河流介导的外源DNA输入可能导致淡水物种被检测到。Yamamoto等(2016)认为淡水密度小于海水, 在海水表面流动可能会将淡水鱼类的eDNA带至沿岸河口区域, 使靠近河流的样点容易在其表层水样中检测到淡水鱼类序列。本研究中, 吻虾虎鱼属以及鲤科各属淡水鱼类物种在一级潮沟尤其是其下游接近内陆河道的样点丰度明显高于其他样点。其次, 养殖池塘以及水产市场排污入河有可能将养殖鱼类的DNA引入采样区域, 从而造成假阳性。有报道称研究区域附近存在罗非鱼(非鲫)、黄颡鱼(Tachysurus fulvidraco)等人工养殖(孙金凤和刘清志, 2010; 蒋万钊和隋凯港, 2015), 推测本研究中检测到的黄颡鱼以及多种非鲫属鱼类序列与此有关。最后, 捕食者粪便中含有的鱼类DNA在进入水体后也有可能会被检测到。研究区域所在的保护区内拥有数量众多的鸟类, 是环西太平洋鸟类迁徙重要的“中转站”、越冬栖息地和繁殖地(陈为峰等, 2003), 鸟类作为鱼类的主要捕食者, 可能在鱼类种群及其DNA扩散过程中扮演着重要角色。因此, 环境DNA技术发现的这28种非黄河口、莱州湾海域鱼类物种, 可能包括一些随机出现在该区域或者种群数量极少的物种, 利用传统的调查方法可能难于发现其踪迹, 而环境DNA技术可以更加灵敏地检测到丰度低、生物量少的稀有物种(Deiner et al, 2017)。
3.2 环境DNA宏条形码技术检测黄河口湿地潮间带鱼类多样性的可行性与影响因素
河口湿地是海陆交替的过渡区域, 具有复杂多样的生境条件, 为河口及近海渔业生物提供了丰富的食物和良好的栖息地(Garwood et al, 2019), 支撑着近海渔业资源的可持续产出。所以, 及时了解河口湿地特别是潮沟系统的鱼类多样性对河口湿地生物多样性保护及邻近水域渔业资源的可持续发展至关重要(马巧珍等, 2023)。本研究通过环境DNA技术在黄河口滨海湿地潮沟中共检测到鱼类55种, 其中黄河口本地鱼类27种, 占比为49.1%, 基本包括了黄河口湿地常见的鱼类物种。国内其他河口湿地也开展了相似的研究并取得了较好的效果。王汝贤等(2023)通过环境DNA技术在长江口检测到鱼类55种, 其中长江口鱼类占比为81.8%; 蒋佩文等(2022)在珠江河口检测到鱼类175种, 其中珠江口鱼类占比65.7%; 李红婷等(2022)在珠江河口伶仃洋海域共检测到鱼类35种; Zou等(2020)在珠江口南沙滨海湿地共检测到鱼类57种。虽然鱼类物种数量和组成在各个研究区域间存在差异, 但环境DNA技术在各个河口湿地鱼类多样性研究中的初步应用表明, 相较传统调查方法的采样难度大、成本高、对研究人员专业素养要求高等, 该技术已展现出一定的优势。
目前, 环境DNA宏条形码技术在河口及近海区域的应用尚处于起步阶段, 而且相比成熟的传统调查手段, 其物种检测效率受到多种因素的影响, 如研究区域的环境特征、采样方法、参考数据库的完善程度、所选引物的物种区分能力等。首先, 较高的温度、太阳辐射和海洋环境会加快eDNA的衰变速率, 这使得对应条件下检测到的eDNA与源生物之间的空间相关性更高(Strickler et al, 2015; Lamb et al, 2022)。因此, 相较淡水环境, 河口湿地咸淡水交汇, 水体沟通频繁, 受潮汐活动影响大, 且滩涂缺少高大植被遮挡, 太阳辐射强烈, 水体深度较浅, 有利于光穿透, 这样的环境特征使eDNA在环境中的存留时间变短, 检测到的eDNA能够更好地反映当下鱼类群落结构的实际面貌, 序列丰度与物种丰度之间相关性更强(Thomsen et al, 2012a; Yamamoto et al, 2017; Jo, 2023)。其次, 过滤水样的体积对于检测到的鱼类物种数有直接影响。环境DNA领域内过滤水样富集eDNA, 单个水样体积通常设置在0.5-2.0 L (Tsuji et al, 2019), 但将不同深度水层的水样混合以及过滤更大体积的水样有利于检测到更多的物种(蒋佩文等, 2022)。另外, 数据库中物种序列信息的完善程度会影响序列比对与分类注释的结果。当库中缺乏相关物种序列信息时, 会比对到序列相似的其他物种, 从而导致结果不准确或只能得到高分类阶元的分类信息; 而当库中比对到的物种信息过多时也会对分类注释造成干扰。因此, 与数据库进行序列比对时, 需要设定相应的筛选标准; 在分析“假阳性”结果时, 需要综合历史调查物种名录与研究区域环境特征谨慎做出判断; 还需要尽早构建本地的条形码数据库, 不仅能够满足土著鱼类多样性的调查需求, 也排除了公共数据库中冗余信息的影响, 提高了环境DNA技术检测的效率(郜星辰和姜伟, 2021)。此外, 引物的物种区分能力以及扩增偏好性也会导致扩增测序过程产生偏差, 可能使测序结果不准确(Knight et al, 2018)。本研究所使用的鱼类经典12S rRNA引物MiFish-E的高变区拥有足够的信息可将鱼类鉴定至不同分类阶元, 其有效性在诸多研究中得到证实(Nakagawa et al, 2018; 蒋佩文等, 2022)。但该引物也存在局限性, 如: 在近似种之间缺乏足够的种间变异, 可能会低估物种丰富度; 很难成功扩增所提取eDNA中包含的全部鱼类信息从而造成假阴性; 偶尔会检测到其他非鱼类物种, 如海鸟、海豚等(Miya et al, 2015, 2020)。研究人员在选择引物时可基于本地物种探索适合的最优遗传标记, 或使用多对引物进行扩增。
3.3 潮沟系统鱼类多样性对水文连通的响应
水文连通可通过影响湿地重要环境因子来影响生物定居、迁移扩散和繁殖等行为, 进而影响湿地的生物连通性以及生物多样性(王新艳等, 2019)。研究区域一级潮沟至三级潮沟流速与断面面积依次递减, 表明水文连通性也依次递减(骆梦等, 2018)。径流量最大的一级潮沟水动力变化复杂, 受潮汐影响作用强烈, 水体交换频繁, 高流速的水体冲刷使其受扰动程度较大, 水文连通强度最高; 三级潮沟水体流速缓慢, 水量小, 受蒸发严重, 易停滞, 有机质富集水平高, 同样受干扰程度较强, 水文连通强度最小; 二级潮沟作为该典型潮沟系统的中间组成部分, 水文连通强度中等(董芮等, 2020)。本研究中, Chao 1指数和ACE指数(两者在生态学中被用于评估群落中的物种总数, 两者的计算公式均赋予了稀有物种一定的计算权重, 但后者对稀有物种定义范围更广, 且校正了变异系数和样本覆盖度)在不同级别潮沟之间呈现出一致的变化趋势。Shannon指数和Simpson指数亦呈现出相同的规律。二级潮沟无论是群落多样性还是丰富度都明显高于其他两级潮沟, 说明其物种种类最多, 并且物种个体数目分配更均匀。这符合中度干扰假说(the intermediate disturbance hypothesis), 即最高的多样性维持在干扰的中间尺度(Connell, 1978)。Sousa (1979)通过研究潮间带受海浪影响的不同大小卵石上定植的物种群落结构也发现中等大小的卵石包含最多样的物种群落。Evangelisti等(2013)对湿地3个不同盐度梯度的沿海水塘进行水样16S rRNA测序, 同样发现微生物群落在中等盐度水平有着最大的多样性。多样性是综合考虑丰富度和均匀度的结果, 三级潮沟群落丰富度小于一级潮沟, 但多样性却大于一级潮沟, 即三级潮沟的均匀度较高。O'Donnell等(2017)发现eDNA测序得到的多样性指数在环境梯度上呈现出线性变化, 随着离岸距离增加, 丰富度下降, 而均匀度上升, 原因是当生物组织颗粒稀少时, 从固定体积的样本中所收集到的量就会更少, 从而降低了检测到更多物种(丰富度)的概率, 并增加了任意两个组织颗粒属于同一物种(均匀度)的可能性。本研究中一级潮沟作为该典型潮沟系统的主潮沟, 上游与海直接相连, 下游与内陆河道相接, 推测多条输入途径使其相较三级潮沟有机会容纳更多物种的eDNA。
研究结果显示优势种矛尾刺虾虎鱼、鮻的序列丰度在水文连通强度大的潮沟中较高, 推测与潮沟生态环境特征不同所导致的环境因子差异有关。秋季是丰水期, 此时水文连通强度大的潮沟径流量大、流速高, 水流会将一定的硅酸盐随泥沙迁走使硅酸盐含量降低(张晓晓等, 2010); 还会冲刷底层大颗粒泥沙, 导致土壤硬度降低, 孔隙大, 渗透性增高, 易被搅动从而吸引甲壳类来此觅食底栖微藻(Yin et al, 2020), 而甲壳类是黄河三角洲矛尾刺虾虎鱼最主要的饵料来源(范海洋等, 2005), 因此, 水文连通强度最大的一级潮沟硅酸盐浓度显著低于三级潮沟, 并且矛尾刺虾虎鱼序列丰度最高。鮻作为广盐性鱼类在低盐环境下可节省用于渗透调节的能量, 并且其幼鱼对低盐度的水流具有强烈的趋流性(李明德等, 1997); 张鼎元等(2020)发现鮻幼鱼在盐度15的条件下有最优的增重率以及最低的饵料系数。与上述研究结论一致, 研究区域一级潮沟的盐度显著低于三级潮沟, 并且随潮沟级别增加呈现出递增的趋势, 而鮻的序列丰度在三级潮沟下游最低, 并已不是优势类群; RDA排序图也表明鮻的序列丰度与盐度呈负相关。
4 结论
本研究使用环境DNA宏条形码技术评估了黄河口典型潮沟系统水体中的鱼类多样性, 共检测出鱼类55种, 其中本地鱼类27种、非本地鱼类28种, 物种组成以鲈形目为主。通过与历史数据对比证实了该技术检测黄河口滨海湿地潮沟鱼类物种组成的可行性与有效性。此外, 研究发现不同水文连通性的潮沟间鱼类物种组成与多样性不同, 二级潮沟群落多样性水平、丰富度、物种种类以及各种鱼类类群中的个体均匀程度等都明显高于其他两级潮沟。6种环境因子硅酸盐(SiO32--Si)、硝酸盐(NO3--N)、pH值、盐度(SAL)、铵盐(NH4+-N)、溶解氧(DO)与鱼类群落结构显著相关。上述结果证实了水文连通性通过改变水文条件会影响鱼类物种组成与分布, 不同级别潮沟之间由于水文连通强度不同会在多种环境理化指标上呈现出显著差异。
虽然环境DNA宏条形码技术仍存在诸多问题, 如对本地数据库的过度依赖、无法获取生物个体生物学指标信息、容易出现假阳性等, 而且, 由于环境DNA衰变机制不具有普遍性, 在不同环境及不同物种中均不相同, 这使得能否用环境DNA浓度及序列丰度表征物种在自然环境中的实际生物量与丰度备受争议。但相比传统的鱼类调查方法, 该技术具有省时省力、灵敏度高、对生物和生态环境无害等诸多优势。随着本地数据库的不断完善及实验流程的标准化, 该技术作为辅助检测手段, 与传统调查方法相结合能更有效地反映鱼类多样性的实际面貌, 为湿地生物多样性监测及资源调查评估提供有力的数据支持。
参考文献
Environmental DNA detection of rare and invasive fish species in two Great Lakes tributaries
DOI:10.1111/mec.14395
PMID:29087006
[本文引用: 1]

The extraction and characterization of DNA from aquatic environmental samples offers an alternative, noninvasive approach for the detection of rare species. Environmental DNA, coupled with PCR and next-generation sequencing ("metabarcoding"), has proven to be very sensitive for the detection of rare aquatic species. Our study used a custom-designed group-specific primer set and next-generation sequencing for the detection of three species at risk (Eastern Sand Darter, Ammocrypta pellucida; Northern Madtom, Noturus stigmosus; and Silver Shiner, Notropis photogenis), one invasive species (Round Goby, Neogobius melanostomus) and an additional 78 native species from two large Great Lakes tributary rivers in southern Ontario, Canada: the Grand River and the Sydenham River. Of 82 fish species detected in both rivers using capture-based and eDNA methods, our eDNA method detected 86.2% and 72.0% of the fish species in the Grand River and the Sydenham River, respectively, which included our four target species. Our analyses also identified significant positive and negative species co-occurrence patterns between our target species and other identified species. Our results demonstrate that eDNA metabarcoding that targets the fish community as well as individual species of interest provides a better understanding of factors affecting the target species spatial distribution in an ecosystem than possible with only target species data. Additionally, eDNA is easily implemented as an initial survey tool, or alongside capture-based methods, for improved mapping of species distribution patterns.© 2017 John Wiley & Sons Ltd.
Crisis of wetlands in the Yellow River Delta and its protection
黄河三角洲湿地面临的问题及其保护
High diversity of trees and corals is maintained only in a nonequilibrium state
DOI:10.1126/science.199.4335.1302
PMID:17840770
[本文引用: 1]

The commonly observed high diversity of trees in tropical rain forests and corals on tropical reefs is a nonequilibrium state which, if not disturbed further, will progress toward a low-diversity equilibrium community. This may not happen if gradual changes in climate favor different species. If equilibrium is reached, a lesser degree of diversity may be sustained by niche diversification or by a compensatory mortality that favors inferior competitors. However, tropical forests and reefs are subject to severe disturbances often enough that equilibrium may never be attained.
The ecologist's field guide to sequence-based identification of biodiversity
DOI:10.1111/mee3.2016.7.issue-9 URL [本文引用: 1]
Ecological effects of wetland hydrological connectivity: Problems and prospects
湿地水文连通的生态效应研究进展及发展趋势
Environmental DNA metabarcoding: Transforming how we survey animal and plant communities
DOI:10.1111/mec.14350
PMID:28921802
[本文引用: 1]

The genomic revolution has fundamentally changed how we survey biodiversity on earth. High-throughput sequencing ("HTS") platforms now enable the rapid sequencing of DNA from diverse kinds of environmental samples (termed "environmental DNA" or "eDNA"). Coupling HTS with our ability to associate sequences from eDNA with a taxonomic name is called "eDNA metabarcoding" and offers a powerful molecular tool capable of noninvasively surveying species richness from many ecosystems. Here, we review the use of eDNA metabarcoding for surveying animal and plant richness, and the challenges in using eDNA approaches to estimate relative abundance. We highlight eDNA applications in freshwater, marine and terrestrial environments, and in this broad context, we distill what is known about the ability of different eDNA sample types to approximate richness in space and across time. We provide guiding questions for study design and discuss the eDNA metabarcoding workflow with a focus on primers and library preparation methods. We additionally discuss important criteria for consideration of bioinformatic filtering of data sets, with recommendations for increasing transparency. Finally, looking to the future, we discuss emerging applications of eDNA metabarcoding in ecology, conservation, invasion biology, biomonitoring, and how eDNA metabarcoding can empower citizen science and biodiversity education.© 2017 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Effects of hydrological connectivity on the community structure of macrobenthos in West Dongting Lake
水文连通性对西洞庭湖大型底栖动物群落结构的影响
The relationship between salinity and bacterioplankton in three relic coastal ponds (Macchiatonda Wetland, Italy)
DOI:10.4236/jwarp.2013.59087 URL [本文引用: 1]
Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding
DOI:10.1111/1755-0998.12433
PMID:26032773
[本文引用: 1]

Freshwater fauna are particularly sensitive to environmental change and disturbance. Management agencies frequently use fish and amphibian biodiversity as indicators of ecosystem health and a way to prioritize and assess management strategies. Traditional aquatic bioassessment that relies on capture of organisms via nets, traps and electrofishing gear typically has low detection probabilities for rare species and can injure individuals of protected species. Our objective was to determine whether environmental DNA (eDNA) sampling and metabarcoding analysis can be used to accurately measure species diversity in aquatic assemblages with differing structures. We manipulated the density and relative abundance of eight fish and one amphibian species in replicated 206-L mesocosms. Environmental DNA was filtered from water samples, and six mitochondrial gene fragments were Illumina-sequenced to measure species diversity in each mesocosm. Metabarcoding detected all nine species in all treatment replicates. Additionally, we found a modest, but positive relationship between species abundance and sequencing read abundance. Our results illustrate the potential for eDNA sampling and metabarcoding approaches to improve quantification of aquatic species diversity in natural environments and point the way towards using eDNA metabarcoding as an index of macrofaunal species abundance.© 2015 The Authors. Molecular Ecology Resources Published by John Wiley & Sons Ltd.
Research of fishery biology of the neritic fish Synechogobius ommaturus in the area of the Huanghe Delta
黄河三角洲斑尾复虾虎鱼渔业生物学的研究
Hydrologic connectivity and the contribution of stream headwaters to ecological integrity at regional scales
DOI:10.1111/j.1752-1688.2007.00002.x URL [本文引用: 1]
The construction and application of BLAST database of DNA barcode for common fish in the Three Gorges Reservoir
三峡库区常见鱼类DNA条形码本地BLAST数据库的构建和应用
Site fidelity and habitat use by young-of-the-year transient fishes in salt marsh intertidal creeks
DOI:10.1007/s12237-019-00576-4
[本文引用: 1]

The extensive use of salt marsh creeks by nekton is widely recognized, yet few investigations have quantified the extent to which tidal migratory nekton make repeated visits to the same location within creek networks. An improved understanding of nekton movements and fidelity can improve insights into the nursery function and value of creek habitats. We conducted tag-recapture experiments with young fishes to determine movements among intertidal creeks in the North Inlet estuary, South Carolina. In 2008, 3246 fishes were tagged with coded microwire tags. In 2012, 577 fishes were tagged with passive integrated transponder (PIT) tags. Based on percentages of recaptures, creek-specific fidelity was determined for spot (Leiostomus xanthurus) (78%), silver perch (Bairdiella chrysoura) (71%), pinfish (Lagodon rhomboides) (80%), and mullets (Mugil curema and Mugil cephalus) (77%). Recaptures occurred up to 79days after release. Rates of recapture of all tagged fishes combined varied among creeks (74% to 100%). Considering earlier research that demonstrated differences in habitat quality among local intertidal creeks, we suggest that differences in fidelity reflect differences in habitat quality among creeks. Greater rates of repeated use (higher fidelity) could help explain why some creeks consistently support more tidal migratory nekton than other creeks. Changes in the strength of fidelity over time could be a useful measure of changes in habitat quality. We suggest that site fidelity be considered as another metric to use in efforts by resource managers to identify and evaluate essential fish habitat.
Consumer-resource coupling in wet-dry tropical rivers
DOI:10.1111/j.1365-2656.2011.01925.x
PMID:22103689
[本文引用: 1]

1. Despite implications for top-down and bottom-up control and the stability of food webs, understanding the links between consumers and their diets remains difficult, particularly in remote tropical locations where food resources are usually abundant and variable and seasonal hydrology produces alternating patterns of connectivity and isolation. 2. We used a large scale survey of freshwater biota from 67 sites in three catchments (Daly River, Northern Territory; Fitzroy River, Western Australia; and the Mitchell River, Queensland) in Australia's wet-dry tropics and analysed stable isotopes of carbon (δ(13) C) to search for broad patterns in resource use by consumers in conjunction with known and measured indices of connectivity, the duration of floodplain inundation, and dietary choices (i.e. stomach contents of fish). 3. Regression analysis of biofilm δ(13) C against consumer δ(13) C, as an indicator of reliance on local food sources (periphyton and detritus), varied depending on taxa and catchment. 4. The carbon isotope ratios of benthic invertebrates were tightly coupled to those of biofilm in all three catchments, suggesting assimilation of local resources by these largely nonmobile taxa. 5. Stable C isotope ratios of fish, however, were less well-linked to those of biofilm and varied by catchment according to hydrological connectivity; the perennially flowing Daly River with a long duration of floodplain inundation showed the least degree of coupling, the seasonally flowing Fitzroy River with an extremely short flood period showed the strongest coupling, and the Mitchell River was intermediate in connectivity, flood duration and consumer-resource coupling. 6. These findings highlight the high mobility of the fish community in these rivers, and how hydrological connectivity between habitats drives patterns of consumer-resource coupling.© 2011 The Authors. Journal of Animal Ecology © 2011 British Ecological Society.
Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method
DOI:10.1016/j.scitotenv.2021.150587 URL [本文引用: 1]
Investigating the fish diversity in Pearl River Estuary based on environmental DNA metabarcoding and bottom trawling
基于环境DNA宏条码和底拖网的珠江河口鱼类多样性
Study on ecological pond aquaculture technology of Tachysurus fulvidraco in saline-alkali land
盐碱地黄颡鱼池塘生态养殖技术研究
Correlation between the number of eDNA particles and species abundance is strengthened by warm temperature: Simulation and meta-analysis
DOI:10.1007/s10750-022-05036-y [本文引用: 1]
Best practices for analysing microbiomes
DOI:10.1038/s41579-018-0029-9
PMID:29795328
[本文引用: 1]

Complex microbial communities shape the dynamics of various environments, ranging from the mammalian gastrointestinal tract to the soil. Advances in DNA sequencing technologies and data analysis have provided drastic improvements in microbiome analyses, for example, in taxonomic resolution, false discovery rate control and other properties, over earlier methods. In this Review, we discuss the best practices for performing a microbiome study, including experimental design, choice of molecular analysis technology, methods for data analysis and the integration of multiple omics data sets. We focus on recent findings that suggest that operational taxonomic unit-based analyses should be replaced with new methods that are based on exact sequence variants, methods for integrating metagenomic and metabolomic data, and issues surrounding compositional data analysis, where advances have been particularly rapid. We note that although some of these approaches are new, it is important to keep sight of the classic issues that arise during experimental design and relate to research reproducibility. We describe how keeping these issues in mind allows researchers to obtain more insight from their microbiome data sets.
Factors structuring estuarine and coastal fish communities across Japan using environmental DNA metabarcoding
DOI:10.1016/j.ecolind.2020.107216 URL [本文引用: 1]
Systematic review and meta-analysis: Water type and temperature affect environmental DNA decay
DOI:10.1111/1755-0998.13627
PMID:35510730
[本文引用: 1]

Environmental DNA (eDNA) has been used in a variety of ecological studies and management applications. The rate at which eDNA decays has been widely studied but at present it is difficult to disentangle study-specific effects from factors that universally affect eDNA degradation. To address this, a systematic review and meta-analysis was conducted on aquatic eDNA studies. Analysis revealed eDNA decayed faster at higher temperatures and in marine environments (as opposed to freshwater). DNA type (mitochondrial or nuclear) and fragment length did not affect eDNA decay rate, although a preference for < 200 bp sequences in the available literature means this relationship was not assessed with longer sequences (e.g. > 800 bp). At present, factors such as ultraviolet light, pH, and microbial load lacked sufficient studies to feature in the meta-analysis. Moving forward, we advocate researching these factors to further refine our understanding of eDNA decay in aquatic environments.This article is protected by copyright. All rights reserved.
Characteristics of carbon and nitrogen distribution in typical tidal creeks of the Yellow River Delta
DOI:10.31497/zrzyxb.20200217 URL [本文引用: 1]
黄河口典型潮沟土壤碳氮分布特征规律
DOI:10.31497/zrzyxb.20200217
[本文引用: 1]

为探究黄河三角洲盐沼土壤碳氮含量在潮沟水系中的时空分布特征,选取黄河口一条典型的潮沟系统,采集一、二、三级潮沟表层土壤,探寻土壤有机碳、总氮与土壤容重、盐度、pH等理化因子的相关关系。结果表明:土壤有机碳和总氮在时空尺度上表现出极大的异质性特征。时间尺度上,土壤有机碳和总氮出现先上升后下降的趋势。空间尺度上,一级潮沟土壤有机碳和总氮平均值(2.9 g·kg<sup>-1</sup>、0.36 g·kg<sup>-1</sup>)大于二级(1.4 g·kg<sup>-1</sup>、0.18 g·kg<sup>-1</sup>)、三级(1.6 g·kg<sup>-1</sup>、0.21 g·kg<sup>-1</sup>)潮沟。相关分析表明,土壤有机碳和总氮与盐度呈显著正相关(P<0.01),与容重呈显著负相关(P<0.01)。盐沼湿地土壤碳氮含量受土壤水盐条件的影响,而潮沟水系的树状结构对水盐条件的影响是导致土壤碳氮含量时空差异分布的重要因素。
Seasonal changes in the community structure of the demersal fishery in Laizhou Bay
DOI:10.3724/SP.J.1118.2013.00137 URL [本文引用: 1]
2010年莱州湾底层渔业生物群落结构及季节变化
Ecological niche of dominant species of fish assemblages in Laizhou Bay, China
莱州湾鱼类群落优势种生态位
Species composition of fishes in the Pearl River Estuary based on environmental DNA metabarcoding
基于环境DNA宏条形码的珠江河口鱼类种类组成
An overview of Chinese coastal wetland and their ecosystem services
中国滨海湿地及其生态系统服务功能研究概述
Hydrological connectivity characteristics and ecological effects of a typical tidal channel system in the Yellow River Delta
黄河三角洲典型潮沟系统水文连通特征及其生态效应
Effects of tidal creek connectivity on fish communities in the Yangtze estuary wetlands
长江口湿地潮沟连通程度对鱼类群落的影响
MiFish metabarcoding: A high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samples
DOI:10.1007/s12562-020-01461-x
[本文引用: 1]

We reviewed the current methodology and practices of the DNA metabarcoding approach using a universal PCR primer pair MiFish, which co-amplifies a short fragment of fish DNA (approx. 170 bp from the mitochondrial 12S rRNA gene) across a wide variety of taxa. This method has mostly been applied to biodiversity monitoring using environmental DNA (eDNA) shed from fish and, coupled with next-generation sequencing technologies, has enabled massively parallel sequencing of several hundred eDNA samples simultaneously. Since the publication of its technical outline in 2015, this method has been widely used in various aquatic environments in and around the six continents, and MiFish primers have demonstrably outperformed other competing primers. Here, we outline the technical progress in this method over the last 5 years and highlight some case studies on marine, freshwater, and estuarine fish communities. Additionally, we discuss various applications of MiFish metabarcoding to non-fish organisms, single-species detection systems, quantitative biodiversity monitoring, and bulk DNA samples other than eDNA. By recognizing the MiFish eDNA metabarcoding strengths and limitations, we argue that this method is useful for ecosystem conservation strategies and the sustainable use of fishery resources in “ecosystem-based fishery management” through continuous biodiversity monitoring at multiple sites.
Use of mitogenomic information in teleostean molecular phylogenetics: A tree-based exploration under the maximum-parsimony optimality criterion
We explored the phylogenetic utility and limits of the individual and concatenated mitochondrial genes for reconstructing the higher-level relationships of teleosts, using the complete (or nearly complete) mitochondrial DNA sequences of eight teleosts (including three newly determined sequences), whose relative phylogenetic positions were noncontroversial. Maximum-parsimony analyses of the nucleotide and amino acid sequences of 13 protein-coding genes from the above eight teleosts, plus two outgroups (bichir and shark), indicated that all of the individual protein-coding genes, with the exception of ND5, failed to recover the expected phylogeny, although unambiguously aligned sequences from 22 concatenated transfer RNA (tRNA) genes (stem regions only) recovered the expected phylogeny successfully with moderate statistical support. The phylogenetic performance of the 13 protein-coding genes in recovering the expected phylogeny was roughly classified into five groups, viz. very good (ND5, ND4, COIII, COI), good (COII, cyt b), medium (ND3, ND2), poor (ND1, ATPase 6), and very poor (ND4L, ND6, ATPase 8). Although the universality of this observation was unclear, analysis of successive concatenation of the 13 protein-coding genes in the same ranking order revealed that the combined data sets comprising nucleotide sequences from the several top-ranked protein-coding genes (no 3rd codon positions) plus the 22 concatenated tRNA genes (stem regions only) best recovered the expected phylogeny, with all internal branches being supported by bootstrap values >90%. We conclude that judicious choice of mitochondrial genes and appropriate data weighting, in conjunction with purposeful taxonomic sampling, are prerequisites for resolving higher-level relationships in teleosts under the maximum-parsimony optimality criterion.Copyright 2000 Academic Press.
MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species
Dispersion and degradation of environmental DNA from caged fish in a marine environment
DOI:10.1007/s12562-018-1282-6
[本文引用: 1]

Environmental DNA (eDNA) consists of DNA fragments shed from organisms into the environment, and can be used to identify species presence and abundance. This study aimed to reveal the dispersion and degradation processes of eDNA in the sea. Caged fish were set off the end of a pier in Maizuru Bay, the Sea of Japan, and their eDNA was traced at sampling stations located at the cage and 10, 30, 100, 300, 600 and 1000m distances from the cage along two transect lines. Sea surface water was collected at each station at 0, 2, 4, 8, 24 and 48h after setting the cage, and again after removing the cage. Quantitative PCR analyses using a species-specific primer and probe set revealed that the target DNA was detectable while the cage was present and for up to 1h after removing the cage, but not at 2h or later. Among the 57 amplified samples, 45 (79%) were collected within 30m from the cage. These results suggest that eDNA can provide a snapshot of organisms present in a coastal marine environment.
Comparing local- and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods
DOI:10.1111/fwb.2018.63.issue-6 URL [本文引用: 1]
Spatial distribution of environmental DNA in a nearshore marine habitat
A review of research advancement on fisheries biology of Japanese Spanish Mackerel Scomberomorus niphonius
蓝点马鲛渔业生物学研究进展
Space-time dynamics in monitoring neotropical fish communities using eDNA metabarcoding
DOI:10.1016/j.scitotenv.2020.142096 URL [本文引用: 1]
Comparison of fish detections, community diversity, and relative abundance using environmental DNA metabarcoding and traditional gears
DOI:10.1002/edn3.v1.4 URL [本文引用: 1]
Biota connect aquatic habitats throughout freshwater ecosystem mosaics
DOI:10.1111/1752-1688.12634
PMID:31296983
[本文引用: 1]

Freshwater ecosystems are linked at various spatial and temporal scales by movements of biota adapted to life in water. We review the literature on movements of aquatic organisms that connect different types of freshwater habitats, focusing on linkages from streams and wetlands to downstream waters. Here, streams, wetlands, rivers, lakes, ponds, and other freshwater habitats are viewed as dynamic freshwater ecosystem mosaics (FEMs) that collectively provide the resources needed to sustain aquatic life. Based on existing evidence, it is clear that biotic linkages throughout FEMs have important consequences for biological integrity and biodiversity. All aquatic organisms move within and among FEM components, but differ in the mode, frequency, distance, and timing of their movements. These movements allow biota to recolonize habitats, avoid inbreeding, escape stressors, locate mates, and acquire resources. Cumulatively, these individual movements connect populations within and among FEMs and contribute to local and regional diversity, resilience to disturbance, and persistence of aquatic species in the face of environmental change. Thus, the biological connections established by movement of biota among streams, wetlands, and downstream waters are critical to the ecological integrity of these systems. Future research will help advance our understanding of the movements that link FEMs and their cumulative effects on downstream waters.
Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system
DOI:10.1016/j.biocon.2016.03.010 URL [本文引用: 1]
Explore the distribution and influencing factors of fish in major rivers in Beijing with eDNA metabarcoding technology
DOI:10.17520/biods.2022240
[本文引用: 1]

Aims: We used eDNA metabarcoding to detect fish diversity in three river systems of Beijing during summer and autumn. The aim of this study is to explore new methods applicable to monitor and protect fish diversity and study the spatio-temporal patterns of fish community in Beijing. Methods: A total of 34 sites were surveyed simultaneously using the eDNA metabarcoding and ground cage methods. The main processes of the eDNA method are water sample collection and enrichment, DNA extraction with DNeasy Tissue and Blood DNA, PCR amplification with prism of MiFish-U, high-throughput sequencing by Illumina Miseq and bioinformatics analysis. The data of eDNA metabarcoding, environmental factors and human activity were used for statistical analysis between fish communities and influencing factors. The main processes of the ground cage with three replications were placed at each site waiting 24 h to be collected for species identification, and the species with number were recorded for subsequent analysis. Results: The result showed that 55 fish species were detected by eDNA metabarcoding was higher than that captured by the ground cage (35 species). The fish community composition was dominated by Cypriniformes and Perciformes. The diversity of clearwater fish in mountain rivers is significantly higher than urban rivers. Urban rivers (the Beiyun River System) were dominated by pollution-tolerant species such as Carassius auratus, Pseudorasbora parva and Misgurnus anguillicaudatus, with a relatively homogeneous community structure. Mountain rivers (Chaobai River System and Daqing River System) are dominated by Zacco platypus, Rhynchocypris lagowskii and Opsariichthys uncirostris. The community structure of clearwater fish was affected by electrical conductivity and total dissolved solids in summer but by altitude and temperature in autumn. The correlation between the abundance of clearwater fish with environmental factors and human activities showed that the abundance of clearwater fish decreased significantly with the increase by turbidity of water and human activities, and was significant correlation with altitude and temperature. Conclusion: The study used eDNA metabarcoding to show the diversity and spatio-temporal patterns of major rivers in Beijing, with a focus on the distribution of clearwater fish. The results demonstrate the feasibility of eDNA metabarcoding for monitoring fish diversity and spatial-temporal distribution in Beijing.
基于eDNA metabarcoding探究北京市主要河流鱼类分布及影响因素
DOI:10.17520/biods.2022240
[本文引用: 1]

使用eDNA宏条形码(eDNA metabarcoding)和地笼法检测了北京市3条水系在夏季和秋季两个季节的鱼类多样性, 旨在研究北京市鱼类群落的空间格局特征, 探索适用于北京鱼类生物多样性监测及保护的新方法。结果表明: 在北京市的34个采样点中, 利用eDNA metabarcoding共检测出鱼类55种, 显著高于传统方法所捕获的鱼类种数(35种), 鱼类组成以鲤形目和鲈形目为主。山区河流清水鱼的多样性要显著高于城区河流, 城区河流(北运河水系)群落结构较为单一, 以鲫(Carassius auratus)、麦穗鱼(Pseudorasbora parva)、泥鳅(Misgurnus anguillicaudatus)等耐污种为优势种; 山区河流(潮白河水系及大清河水系)以宽鳍鱲(Zacco platypus)、拉氏鱥(Rhynchocypris lagowskii)、马口鱼(Opsariichthys uncirostris)等为优势种。不同季节影响清水鱼群落结构的环境因子不同, 夏季主要是总溶解固体和电导率, 秋季主要是海拔和温度。清水鱼丰富度与环境因子及人类活动的相关性表明, 清水鱼的丰富度随着总溶解固体及灯光指数增加而显著降低, 且均与海拔、温度等存在显著相关性。本研究证明了eDNA metabarcoding方法用于监测北京市鱼类多样性及其时空分布的可行性。
Disturbance in marine intertidal boulder fields: The nonequilibrium maintenance of species diversity
DOI:10.2307/1936969 URL [本文引用: 1]
Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms
DOI:10.1016/j.biocon.2014.11.038 URL [本文引用: 1]
Research on comprehensive development and utilization of geothermal resources in Dongying
东营市地热资源综合开发与利用研究
Seasonal variations in fish community structure in the Laizhou Bay and the Yellow River Estuary
莱州湾及黄河口水域鱼类群落结构的季节变化
Actualities, problems and suggestions of wetland protection and restoration in the Yellow River Delta
黄河三角洲湿地保护与恢复的现状、问题与建议
Detection of a diverse marine fish fauna using environmental DNA from seawater samples
Monitoring endangered freshwater biodiversity using environmental DNA
DOI:10.1111/j.1365-294X.2011.05418.x URL [本文引用: 1]
The detection of aquatic macroorganisms using environmental DNA analysis—A review of methods for collection, extraction, and detection
DOI:10.1002/edn3.2019.1.issue-2 URL [本文引用: 1]
Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding
DOI:10.1111/mec.13428
PMID:26479867
[本文引用: 1]

Global biodiversity in freshwater and the oceans is declining at high rates. Reliable tools for assessing and monitoring aquatic biodiversity, especially for rare and secretive species, are important for efficient and timely management. Recent advances in DNA sequencing have provided a new tool for species detection from DNA present in the environment. In this study, we tested whether an environmental DNA (eDNA) metabarcoding approach, using water samples, can be used for addressing significant questions in ecology and conservation. Two key aquatic vertebrate groups were targeted: amphibians and bony fish. The reliability of this method was cautiously validated in silico, in vitro and in situ. When compared with traditional surveys or historical data, eDNA metabarcoding showed a much better detection probability overall. For amphibians, the detection probability with eDNA metabarcoding was 0.97 (CI = 0.90-0.99) vs. 0.58 (CI = 0.50-0.63) for traditional surveys. For fish, in 89% of the studied sites, the number of taxa detected using the eDNA metabarcoding approach was higher or identical to the number detected using traditional methods. We argue that the proposed DNA-based approach has the potential to become the next-generation tool for ecological studies and standardized biodiversity monitoring in a wide range of aquatic ecosystems.© 2015 John Wiley & Sons Ltd.
Investigation of biodiversity from spring catch in coastal waters of Laizhou Bay and Huanghe Estuary
莱州湾、黄河口水域春季近岸渔获生物多样性特征的调查研究
Application of environmental DNA technology in fish diversity analysis in the Yangtze River estuary
环境DNA技术在长江口鱼类多样性分析中的应用
Influence of hydrological connectivity of coastal wetland on the biological connectivity of macrobenthos in the Yellow River Estuary
DOI:10.31497/zrzyxb.20191205 URL [本文引用: 1]
黄河口滨海湿地水文连通对大型底栖动物生物连通的影响
DOI:10.31497/zrzyxb.20191205
[本文引用: 1]

滨海湿地处于海陆的交错地带,在人类活动和气候变化的双重作用下导致湿地水文连通受阻。选取黄河口滨海湿地三条水文连通强度差异明显的潮沟,调查不同潮沟的大型底栖动物群落,分析水文连通强度对大型底栖动物分布和生物多样性的影响。结果表明:记录到的大型底栖动物共有52种,优势类群为多毛类和软体动物;总体来看随着水文连通强度的增强,大型底栖动物的总密度呈上升趋势;中等水文连通强度的区域,生物多样性较高;大型底栖动物不同类群的数量和分布对水文连通引起的环境因子变化的响应也有很大的差异,多毛类和寡毛类在水文连通弱的区域占优势,软体动物在水文连通较强营养物质含量丰富的区域占优势,甲壳类在靠近潮沟源头水文连通最强的区域占优势。
Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea
DOI:10.1038/srep40368
PMID:28079122
[本文引用: 1]

Environmental DNA (eDNA) metabarcoding has emerged as a potentially powerful tool to assess aquatic community structures. However, the method has hitherto lacked field tests that evaluate its effectiveness and practical properties as a biodiversity monitoring tool. Here, we evaluated the ability of eDNA metabarcoding to reveal fish community structures in species-rich coastal waters. High-performance fish-universal primers and systematic spatial water sampling at 47 stations covering similar to 11 km(2) revealed the fish community structure at a species resolution. The eDNA metabarcoding based on a 6-h collection of water samples detected 128 fish species, of which 62.5% (40 species) were also observed by underwater visual censuses conducted over a 14-year period. This method also detected other local fishes (>= 23 species) that were not observed by the visual censuses. These eDNA metabarcoding features will enhance marine ecosystem-related research, and the method will potentially become a standard tool for surveying fish communities.
Environmental DNA as a ‘snapshot' of fish distribution: A case study of Japanese Jack Mackerel in Maizuru Bay
Hydrological connectivity and herbivores control the autochthonous producers of coastal salt marshes
DOI:10.1016/j.marpolbul.2020.111638 URL [本文引用: 1]
Linking metrics of landscape pattern to hydrological process in a lotic wetland
DOI:10.1007/s10980-015-0219-z URL [本文引用: 1]
High sensitivity of 454 pyrosequencing for detection of rare species in aquatic communities
DOI:10.1111/2041-210X.12037 URL [本文引用: 1]
Effects of long time low salinity on the growth and physiology in the juvenile of Liza haematocheila
长期低盐胁迫对梭鱼生长及生理的影响
Seasonal variation and fluxes of nutrients in the lower reaches of the Yellow River
黄河下游营养盐浓度季节变化及其入海通量研究
Actualities of wetland hydrological connectivity blocked in the Yellow River Delta
黄河三角洲湿地水文连通受阻现状
eDNA metabarcoding as a promising conservation tool for monitoring fish diversity in a coastal wetland of the Pearl River Estuary compared to bottom trawling
DOI:10.1016/j.scitotenv.2019.134704 URL [本文引用: 1]