DNA条形码(DNA barcode)是基因组中普遍存在的、较短的、标准化的基因序列。由于该DNA序列具有足够的变异程度以区分不同的物种, 因此对其进行分析可实现物种的有效鉴定(Hebert et al, 2003; Taberlet et al, 2007)。自该技术被提出以来, 已经成为了动植物保护、物种鉴定、生物多样性评估、种间互作研究的有力手段之一(Kress et al, 2015)。DNA条形码的序列一般较短(Hebert et al, 2004b), 因此对样品的DNA质量要求较低, 不仅能根据标本或生物体的残骸准确识别物种(Meusnier et al, 2008; Alacs et al, 2010), 还能从环境遗留物中(如动物的毛发、粪便或尿液)鉴别物种痕迹。这种对野生动物不捕捉、低干扰的非损伤性取样(non-invasive sampling)方法对于难以监测或濒临灭绝的物种十分必要(李明等, 2001; Hebert et al, 2004a; Waits & Paetkau, 2005; Valentini et al, 2009)。
脊椎动物DNA条形码研究通常利用线粒体DNA来鉴别物种, 这主要是因为线粒体基因组较核基因组构成简单, 不存在高度变异的内含子, 重组现象较少, 具有足够的种间变异、而种内变异相对稳定, 拷贝数多, 易被扩增(Carracedo et al, 2000; 魏辅文等, 2001; Taberlet et al, 2007; Kress et al, 2015)。早期标准的条形码是编码细胞色素c氧化酶I (COI)基因上600-800 bp的片段(Hebert et al, 2003)。随着测序技术的改进, 科研人员可以对每个主要的分类类群有针对性地设计线粒体DNA靶标序列的通用引物, 从而提高物种鉴别的精确性(Taberlet et al, 1999; 魏辅文等, 2001; Valentini et al, 2009; 俞可心等, 2022)。
中国地域辽阔, 生境类型复杂多样, 是世界上生物多样性最丰富的国家之一(吴慧等, 2022)。四川王朗国家级自然保护区成立于1963年, 是我国建立最早的4个以保护大熊猫等珍稀野生动物及其栖息地为主的自然保护区之一(胡锦矗等, 2011)。保护区面积32,297 ha, 海拔跨度大, 植被类型多样, 动物资源丰富(詹祥江, 2006)。先前的工作中, 保护区管理部门通过样线调查和红外相机陷阱法等方法对保护区内物种进行了长期的野外调查和监测(Li et al, 2012a), 共记录鸟类16目273种(李桂垣和张清茂, 1989; 李桂垣等, 1993; 刘少英等, 2001; 顾人和, 2005; 于笑云, 2009①(①于笑云 (2009) 王朗国家级自然保护区生态旅游市场营销研究. 硕士学位论文, 北京林业大学, 北京.); 尚晓彤等, 2020), 兽类7目77种(魏孝荣, 1988, 1991; 张国修等, 1991; Li et al, 2010, 2012a, b; 段丽娟, 2014; 赵联军等, 2016; 田成等, 2018; 刘少英等, 2019), 两栖类2目9种(刘少英等, 2001; 叶昌媛和费梁, 2007; 李成等, 2017; Zhang et al, 2018), 爬行类2目13种(刘少英等, 2001; 康东伟等, 2010), 对脊椎动物的分布和丰度等有较充分的了解, 对其活动规律也有一定掌握。但传统的监测方法在物种的监测和鉴定上可能存在以下问题: (1)样线法调查中对物种的鉴定主要依据动物的活动痕迹, 可能判断不准确, 如Li等(2012a)发现了调查人员将川金丝猴(Rhinopithecus roxellana)的食迹误判为复齿鼯鼠(Trogopterus xanthipes); (2)围栏陷阱法、铗夜法调查可能造成监测动物死亡, 对物种种群有一定影响; (3)形态鉴定结果和研究者的经验有很大的关系, 可能存在鉴定有误的情况, 如李成等(2017)的研究中通过形态学鉴定认为记录到了高原林蛙(Rana kukunoris), 但在Zhang等(2018)的研究中认为保护区内仅有1种林蛙, 为中国林蛙(Rana chensinensis), 再如秦岭鼢鼠(Eospalax rufescens)在多个研究中的形态学鉴定有不同的结果(赵联军等, 2016), 有待使用遗传学方法进一步分析; 刘少英等(2019)在啮齿目动物的形态学鉴定基础上使用分子系统学方法才证实了该种在四川王朗国家级自然保护区的分布, 说明遗传学方法在物种鉴定方面较形态学方法更能提供可信的参考(Hebert et al, 2003; Valentini et al, 2009)。
掌握保护区现有物种遗传信息, 建立和完善遗传资源库, 是维持生物多样性和生态系统功能的重要手段之一, 但传统监测方法无法获得陆生脊椎动物遗传资源信息。借助分子生物学技术手段不仅可以验证物种鉴定的准确性, 还可以对遗传资源进行本底调查、整理和综合利用。因此, 如何建立陆生脊椎动物DNA条形码数据库, 将生态学和保护遗传学手段相结合, 形成科学有效的保护管理方案, 是保护管理部门亟待解决的问题。
结合野外调查监测和分子遗传学技术, 本研究旨在: (1)建立四川王朗国家级自然保护区陆生脊椎动物DNA条形码数据库, 为陆生脊椎动物的生物多样性系统评估提供基础遗传学数据; (2)初步探讨不同本底调查手段的物种监测效果, 为保护区的保护管理提供重要支撑。
1 研究区域概况
四川王朗国家级自然保护区位于四川省平武县(103°55′-104°10′ E, 32°49′-33°02′ N), 地处青藏高原东缘、岷山山系中段, 属丹巴-松潘半湿润气候, 是全球生物多样性热点区(Myer et al, 2000), 海拔范围2,400-4,900 m (段利娟, 2014②(②段利娟 (2014) 王朗自然保护区大熊猫及其同域物种活动节律及栖息地利用研究. 硕士学位论文, 北京林业大学, 北京.); 陈星等, 2019; Tian et al, 2019), 气候与植被具有明显的垂直分布规律(杨宏伟, 2013③ (③杨宏伟 (2013) 人工林作为大熊猫栖息地适宜性研究. 硕士学位论文, 北京林业大学, 北京.); 尚晓彤等, 2020; 曹亚珍等, 2022)。该保护区是中国最早建立的森林生态系统保护区之一, 是以保护大熊猫(Ailuropoda melanoleuca)、川金丝猴、四川羚牛(Budorcas tibetanus)、绿尾虹雉(Lophophorus lhuysii)等国家一级重点保护野生动物为目标的自然保护区, 除此之外还有许多栖息于此的濒危物种受到保护(田成等, 2018)。
2 研究方法
2.1 样品采集与保存
2020年5月至2021年5月, 根据研究区域地形和生境特征及潜在物种的生活习性, 组织保护区监测骨干定期开展本底调查, 在24条监测路线的基础上设计针对性强的调查样线共31条, 样线长度3-5 km不等, 海拔范围2,400-3,800 m (图1)。由调查人员沿样线以5 km/h的速度行进并搜索, 对可见范围内(约50 m) 新鲜的粪便、尸体、卵壳以及脱落的羽毛和毛发进行采集。采集时先填写样品信息单, 包括样品编号、动物类别、采集点坐标等信息, 然后佩戴好一次性乳胶手套、使用75%乙醇喷雾消毒, 用无菌的镊子、剪刀或棉签, 采集少量深层未腐烂的组织、新鲜的粪便、卵壳, 对于不同个体更换采样工具, 每份样品保存于无菌的离心管中, 并在离心管壁写好标记, 一式三份, 用两倍以上体积95%-100%乙醇浸泡并于-20℃冻存(张保卫等, 2004; 单磊等, 2018), 离心管口用封口膜密封, 避免渗漏、挤压; 羽毛和毛发需保护好根部, 存放在干燥信封中, 常温条件下封存(Segelbacher, 2002)。同时, 根据相关文献资料, 对于栖息在保护区内、但野外调查没有获得样品的物种, 通过采集博物馆(唐家河岷山山系动植物博物馆和四川大学自然博物馆)标本样品作为补充, 用于不同类型样品DNA提取方法、PCR实验条件、引物适用性分析等。
图1
图1
四川王朗国家级自然保护区研究区域及调查样线分布
Fig. 1
The study area and transect line distribution in Wanglang National Nature Reserve (WLNNR)
2.2 样品处理与鉴定
2.2.1 DNA提取、扩增
组织、蛋壳、羽毛及标本样品使用Qiagen试剂盒(DNeasy Blood & Tissue Kit; QIAamp Fast DNA Stool Kit)并参照试剂盒指导步骤进行DNA提取, 其中博物馆标本样品需先经乙醇和灭菌水洗涤2-3次后按照试剂盒步骤进行提取实验; 粪便样品使用QIAamp Fast DNA Stool Kit试剂盒处理。利用分光光度计(NanoDrop2000, Thermo SCIENTIFIC)检测DNA浓度和纯度。通过文献调研确定各类动物通用引物, 如适用于鸟类的通用引物COI-K_ Bird_F/R、L6697Bird_F/H7390Thrush_R等, 兽类通用引物16SF/R及有蹄类通用引物COIung、鼠类通用引物BatL/R6036R等(表1)。对检测合格的DNA样品进行PCR扩增, 产物用1%琼脂糖凝胶120 V电泳检测。
表1 用于PCR扩增和DNA测序的引物
Table 1
| 类别 Category | 目标片段所在基因(引物名称) Target gene (primers' names) | 上游引物序列5'-3' Forward primer sequence 5'-3' | 下游引物序列5'-3' Reverse primer sequence 5'-3' | 退火温度 Annealing temperature (℃) | 循环 次数 Cycles | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 兽类 Mammals | COI (COIung) | GTACCGCTAATAATTGGTGCTCC | GGGTGGCCAAAGAATCAGAACAAGTG | 56 | 35 | Kumar et al, 2017 |
| 16S (16SF/R) | GAGAAGACCCTATGGAGC | ATAGAAACCGACCTGGAT | 50 | 30 | Xiong et al, 2016 | |
| COI (BatL/R6036R) | CCTACTCRGCCATTTTACCTATG | ATCTCTGGGTGTCCAAAGAATCA | 55 | 35 | Robins et al, 2007 | |
| ATP6 (ATP6-DF3/ATP6-DR2) | AACGAAAATCTATTCGCCTCT | TGGATGGACAGTATTTGTTTTGAT | 60/50 | 10/30 | Chaves et al, 2012 | |
| D-loop (BEDL225/H470) | ATGTACATACTGTGCTTGGC | GTCATTAGTCCATCGAGATG | 53 | 38 | Zhang et al, 2002 | |
| 鸟类 Avians | COI (COI-K_Bird_F/R) | CCCCAGACATAGCATTYCC | TTGTGATAGTGGTGGGGTTTTAT | 46/53 | 5/45 | Parejo-Farnés et al, 2018 |
| Cytb (Cytb-L14816/H15173) | CCATCCAACATCTCAGCATGATGAAA | CCCCTCAGAATGATATTTGTCCTCA | 50 | 30 | Parson et al, 2000 | |
| COI (L6697Bird_F/H7390Thrush_R) | TCAACYAACCACAAAGAYATCGGYAC | ACGTGGGARATRATTCCAAATCCTG | 48/51 | 5/30 | Saitoh et al, 2015 | |
| COI (PasserF/R) | CCAACCACAAAGACATCGGAACC | GTAAACTTCTGGGTGACCAAAGAATC | 58 | 35 | Lohman et al, 2009 | |
| 两栖类 Amphibians | COI (LCO1490/HCO2198) | GGTCAACAAATCATAAAGATATTGG | TAAACTTCAGGGTGACCATCA | 50 | 35 | Folmer et al, 1994 |
| COI (LepF/R) | ATTCAACCAATCATAAAGATATTGG | TAAACTTCTGGATGTCCATCA | 46 | 35 | Hebert et al, 2004a | |
| 16S (P7/8) | CGCCTGTTTACCCAT | CCGGTCTGAACTCAGATCACGT | 55 | 36 | Simon et al, 1994 | |
| 16S (Sar/Sbr) | CGCCTGTTTATCCAT | CCGGTCTGAACTCAGATCACGT | 48 | 40 | Palumbi et al, 2002 | |
| 12S-16S (1602L/2571H) | GTATACCGGAAGGTGTACTTGGAACAG | TACCTTCGCACGGTCAGAATACCGC | 50 | 35 | Fu et al, 2007 | |
| Cytb (FrogGlu-f/Thr-r) | TGATCTGCCACCGTTG | CTCCATTCTTCGRCTTACAAG | 46 | 40 | Hillis et al, 1996 | |
| RAG1 (RAG1-F/R) | AGCTGCAGYCARTACCAYAARATGTA | GCAAAGTTTCCGTTCATTCTCAT | 50 | 35 | Fu et al, 2007 |
2.2.2 数据处理与物种鉴定
PCR产物由北京擎科生物科技有限公司使用ABI 3730测序仪进行序列测定, 利用Chromas V2.6.5 (Technelysium Pty Ltd, Australia)查看测序峰图, 并对目标序列进行人工校正, 进而在NCBI (National Center for Biotechnology Information,
通用引物较为保守, 对各类群物种分辨率不同(Hebert et al, 2003; 邵昕宁等, 2019), 通过单一引物扩增后得到的物种信息可能会出现其地理分布与采样地不符, 或与形态学鉴定不符的情况。对此类样品, 本研究采用两对及以上引物进行扩增, 并以多个目标片段比对到同一个物种为判定依据来进行综合研判。参考历史文献, 本研究共选取了16对通用引物, 其中兽类5对, 鸟类4对, 两栖类7对(表1)。对于扩增后浓度过低、无条带等不合格样品, 本研究优化PCR条件, 二次扩增后送测。对于测序结果为双峰的样品, 使用StarPrep PCR和DNA片段纯化试剂盒(GenStar货号D206-01)纯化PCR产物, 利用T-A克隆(TaKaRa, pMDTM-19T Vector)得到目标片段并送测。
对于因NCBI数据库物种信息及序列不够完整而无法确定物种的样品, 根据比对结果中所建议的邻近物种信息, 再对照保护区物种名录, 通过形态学初步判断物种。使用MEGA 11.0 (Tamura et al, 2021)软件的Clustal W功能, 将人工校对后的序列与邻近物种已发表的序列分别对齐并剪切成目标片段长度, 使用最大简约法(maximum parsimony, MP)基于进化树对分重接模型(tree-bisection-reconnection, TBR)构建系统发育树(Sourdis & Nei, 1988), 选择该类群之外亲缘关系最近的物种作为外群, 并对各分支bootstrap置信区间进行1,000次的重复检验。对于单一基因片段区分能力相对较差的物种, 采用两个或以上基因片段拼接进行分析(Fu et al, 2007; Rao & Wilkinson, 2008)。
3 结果
3.1 样品组成
调查队于2020年5月至2021年5月在四川王朗国家级自然保护区总共布设监测样线31条, 覆盖面积约120 km2, 占保护区总面积的37.2%, 共收集样品224份。另从唐家河岷山山系动植物博物馆的32份目标馆藏标本中获取遗留的肌肉组织, 据博物馆名录描述包括14目21科32种; 从四川大学自然博物馆在平武地区(小河沟保护区)采集的样品中获取肌肉组织, 共58份, 样品的物种信息仅记载为鼠类。
本研究对收集的314份样品进行分析, 包括: 鸟类样品156份, 兽类137份, 两栖类21份; 123个粪便, 60个羽毛(毛发), 37个组织, 90个标本遗留肌肉组织, 4个卵壳(表2)。根据个体形态学和样品特征初步判断出物种的样品共161份, 其中具有较高辨识度的主要为大熊猫、毛冠鹿(Elaphodus cephalophus)、亚洲黑熊(Ursus thibetanus)、蓝马鸡(Crossoptilon auritum)等; 难以判断物种来源的主要为羽毛(毛发)、粪便样品, 其中鸟类78份、兽类75份。
表2 样品类别、类型及其数量
Table 2
| 类别 Category | 数量 No. of samples | 样品类型 Sample type | 数量 No. of samples |
|---|---|---|---|
| 鸟类 Avians | 156 | 粪便 Feces | 123 |
| 兽类 Mammals | 137 | 羽毛(毛发) Feathers (Pelage) | 60 |
| 两栖类 Amphibians | 21 | 组织 Tissues | 37 |
| 标本组织 Tissues from specimens | 90 | ||
| 卵壳 Eggshells | 4 | ||
| 总计 Total | 314 | 总计 Total | 314 |
3.2 引物适用性
表3 样品经引物扩增的成功率、识别率
Table 3
| 物种类型(样品数量/个) Category (No. of samples) | 成功扩增样品数(扩增成功率) No. of successfully amplified samples (Success rate) | 通用引物鉴定物种数(识别率) No. of species identified by universal primers (Identification rate) | 特异性引物鉴定物种数(识别率) No. of species identified by specific primers (Identification rate) |
|---|---|---|---|
| 鸟类 Avians (156) | 128 (82.1%) | 41 (89.1%) | 5 (10.9%) |
| 兽类 Mammals (137) | 111 (81.0%) | 15 (55.6%) | 12 (44.4%) |
| 两栖类 Amphibians (21) | 21 (100%) | 3 (75.0%) | - |
3.3 齿突蟾属物种系统发育关系
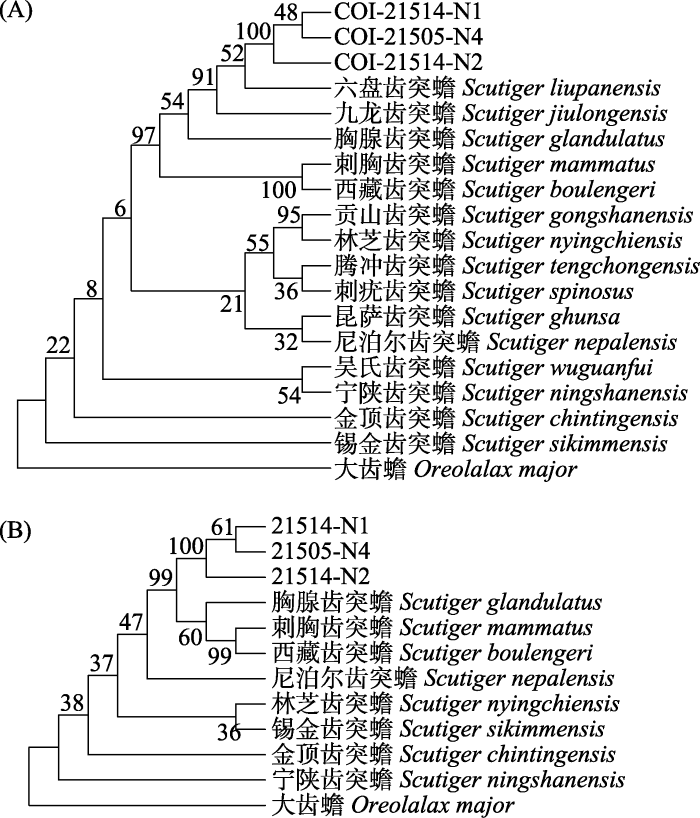
经过拼接、同属物种序列比对、手工校正以及去除低质量片段, 本研究获得了3个齿突蟾样品的线粒体COI、12S-16S、16S、Cytb及核基因RAG1的基因片段。我们将COI基因片段, 以及COI、16S、Cytb 3个基因的拼接片段, 分别与GenBank中下载的8个齿突蟾属物种的序列, 包括: 刺胸齿突蟾(Scutiger mammatus)、西藏齿突蟾(S. boulengeri)、宁陕齿突蟾(S. ningshanensis)、林芝齿突蟾(S. nyingchiensis)、尼泊尔齿突蟾(S. nepalensis)、胸腺齿突蟾(S. glandulatus)、金顶齿突蟾(S. chintingensis)、锡金齿突蟾(S. sikimmensis), 以及外群物种大齿蟾(Oreolalax major)的序列构建系统发育树(图2)。在MP聚类关系中, 部分分支的支持值较低(支持率 < 70%)。
图2
图2
基于COI序列(A)和1,781 bp的COI、16S、Cytb拼接序列(B)构建的齿突蟾属系统发育树。各分支上的数字为分支支持值。
Fig. 2
Phylogenetic tree based on COI sequences of Scutiger (A) and 1,781 bp sequences (COI, 16S, Cytb) of Scutiger (B). Bootstrap values of each branch were showed.
3.4 物种鉴定
通过与NCBI数据库中参考序列的比对, 本研究共鉴定出陆生脊椎动物18目35科74种, 包括: 鸟类10目20科46种, 兽类6目11科24种, 两栖类2目4科4种。其中, 224份四川王朗国家级自然保护区野外采集的样品经提取DNA后, 成功扩增175份(粪便78份, 羽毛及毛发57份, 组织36份, 卵壳4份), 从中共鉴定出14目29科57种, 包含36种鸟类, 17种兽类, 4种两栖类(附录1)。90份来自博物馆的样品鉴定出10种鸟类、7种兽类(附录2)。所有从四川大学自然博物馆取得的样品(58份)均能够得到单一、清晰的PCR扩增条带, 共鉴定出2目2科6种, 均为小型兽类; 唐家河博物馆的样品(32份)中, 成功PCR扩增的为18份, 鉴定出8目10科11种, 包含10种鸟类, 1种兽类。对于缺少参考序列无法判断物种归属的齿突蟾属物种, 结合影像资料中形态学特征和保护区物种名录的记载判定为王朗齿突蟾(Scutiger wanglangensis) (叶昌媛和费梁, 2007)。
表4 物种分子鉴定结果及经验判断正确率
Table 4
| 物种类别 Category | 样品数 No. of samples | 物种预判样品数 No. of morphologically identified samples | 预判正确样 品数 No. of correctly identified samples | 预判正确率 Accuracy of morphological identification rate |
|---|---|---|---|---|
| 鸟类 Avians | 109 | 57 | 30 | 52.6% |
| 兽类 Mammals | 99 | 38 | 18 | 47.4% |
| 两栖类 Amphibians | 13 | 13 | 9 | 69.2% |
| 合计 Total | 221 | 108 | 57 | 52.8% |
4 讨论
本研究收集了来自四川王朗国家级自然保护区及博物馆标本样品, 经DNA提取、PCR扩增、序列比对系列流程进行了物种鉴定, 最终筛选出了16对DNA条形码引物, 共鉴定出陆生脊椎动物18目35科74种(鸟类46种、兽类24种、两栖类4种)的遗传信息。在四川王朗国家级自然保护区调查中所获得的224份样品中, 最终鉴定出陆生脊椎动物14目29科57种, 包括兽类17种、鸟类36种、两栖类4种(附录1), 初步建立了四川王朗国家级自然保护区陆生脊椎动物遗传资源数据库。
本研究中, 四川大学的博物馆标本样品为浸制标本, 采集于2019年, 而唐家河博物馆标本样品为干制标本, 采集时间较久远且信息不完整(仅有部分样品记录为1974年至2002年间采集)。前者全部成功扩增, 后者扩增成功率超过50%, 说明博物馆标本可以用于物种鉴定和遗传资源库建设, 对于保存时间较长的干制标本需要进一步优化提取方法。同时, 本研究选择的通用引物和特异性引物在这些博物馆标本样品中同样适用。
DNA条形码作为物种鉴定的一种方法, 被广泛应用于生物多样性评估和保护研究中, 其不受物种发育阶段、性别及个体完整性的影响, 在调查人员缺少生物学分类经验的情况下还能弥补人为判断的不足(邵昕宁等, 2019)。本研究所采集样品大多来自非损伤性取样, 相较于伤害性取样(destructive sampling)、非伤害性取样(non-destructive sampling)不会对保护区的野生动物造成损伤, 对其活动干扰极低。对非损伤性样品来说, DNA条形码法是对其进行物种鉴定的最有效的手段之一(Parejo-Farnés et al, 2018)。在保护区以往的调查和监测中, 对所采集的非损伤性样品的来源物种仅根据调查人员的经验判断, 往往仅能对部分形态特征独特的样品进行判断, 且很容易出错。例如, 肉食性动物的粪便一般形状、颜色相近, 本研究采集的14份粪便样品中判断错误的有8份, 其中3份黄喉貂(Martes flavigula)的粪便样品被误判为豹猫(Prionailurus bengalensis)和黄鼬(Mustela sibirica); 保护区重点保护动物四川羚牛的两份粪便样品均被错误判断为野猪(Sus scrofa)粪便, 说明利用DNA条形码法进行准确的物种鉴定十分必要。
本研究首次获得了王朗齿突蟾的线粒体遗传信息, 基于最大简约法建立的王朗齿突蟾与其邻近物种的系统发育关系表明, 我们所使用的两栖类通用引物仅能将所获得样品鉴定到齿突蟾属, 未能有效地区分同属不同物种。其主要原因在于: 一方面, 齿突蟾属内部亲缘关系较近(郑渝池等, 2004; Pyron & Wiens, 2011; Hofmann et al, 2017; Dufresnes & Litvinchuk, 2022), 各种间存在频繁的线粒体基因交流(Chen et al, 2009), 仅通过几个线粒体基因片段可能难以解析该属的系统发育关系; 另一方面, 王朗齿突蟾在2007年由叶昌媛和费梁从西藏齿突蟾分出定为新种(叶昌媛和费梁, 2007), 此后没有更进一步的研究, 特别是缺乏分子生物学的研究数据(Dufresnes & Litvinchuk, 2022)。使用更为合适的基因片段或测定全基因组信息有望全面回答齿突蟾属的进化历史问题(车静等, 2000; Hou et al, 2020)。因此, 对于DNA条形码难以清晰区分的物种, 一方面应在DNA条形码的设计上考虑多基因片段以进行综合比对, 对特别重要的物种建议测定全基因组信息作为遗传资源库的重要数据组成。另一方面形态学特征仍是值得参考的佐证, 应结合红外相机等方法获得动物影像资料, 以综合研判物种信息。
在粪便样品中, 基于物种条形码的序列分析除了可以确定样品的物种信息, 还能获得其食物组成的物种信息。前人研究认为, 具有广食谱的捕食性动物可以作为潜在的群落生物多样性的“采样器” (Shao et al, 2021a, b), 对其粪便采取条形码分析是一种可行的生物多样性监测手段。在我们的研究中, 对粪便样品的物种鉴定结果充分说明了这一点。我们对食肉动物(豹猫、黄喉貂、黄鼬)的粪便使用多种引物分别扩增, 在一个黄喉貂粪便中检出了其猎物的DNA片段, 根据序列比对结果和物种分布记录推测为红白鼯鼠Petaurista alborufus, 说明基于DNA条形码技术可以掌握目标物种的食物组成情况, 探究动物物种间的捕食关系, 有效地服务动物群落监测。
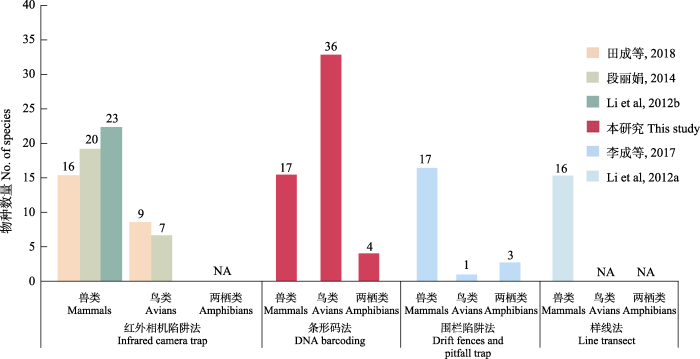
四川王朗国家级自然保护区自成立以来, 开展了长期的野外调查和监测, 常规的物种多样性监测手段主要为红外相机陷阱法、样线法、围栏陷阱法等, 其中红外相机陷阱法是保护区调查本底物种多样性最常用的方法, 该方法主要记录动物活动影像, 根据形态学特征鉴定物种, 已经获得很多历史数据(李晟等, 2020; 田佳等, 2021)。我们对基于DNA条形码技术和其他监测手段所获得的保护区物种多样性数据进行了比较分析。在前人对红外相机数据的分析中, 累计记录到陆生脊椎动物10目37科104种, 包括: 鸟类5目22科77种, 兽类5目15科27种(Li et al, 2010, 2012a, b; 段丽娟, 2014; 田成等, 2018; 尚晓彤等, 2020)。其中, 在同样为期一年的多项调查工作中, 通过红外相机记录到兽类物种分别为16种(田成等, 2018)、20种(段丽娟, 2014)、23种(Li et al, 2012b), 我们通过DNA条形码技术鉴定到兽类17种, 两种方法监测效果相当; 通过红外相机监测到的鸟类物种数分别为9种(田成等, 2018)和7种(段丽娟, 2014), 仅占本研究鉴定出物种数的25.0%和19.4%, 本研究所采用方法在鸟类监测方面具有明显优势(图3)。在两栖类动物调查中, 由于红外相机不适用于监测体温接近环境温度的两栖类动物(Rovero et al, 2010), 所以缺乏相关数据(图3)。因此, 在相同工作时长内, DNA条形码法具有更高的工作效率, 我们通过DNA条形码法监测到的物种总数多于红外相机陷阱法, 且更容易收集到小型动物如鸟类、两栖类动物遗传信息。
图3
图3
动物多样性监测方法的比较(NA为缺乏相应数据)
Fig. 3
Comparison of animal species diversity monitoring methods (NA means relevant data deficiency)
除红外相机陷阱法外, 样线法、围栏陷阱法也是保护区采取的调查和监测手段, 样线法依靠调查人员的经验来判断物种, 更适用于大型兽类监测; 围栏陷阱法主要针对小型地栖脊椎动物, 例如小型兽类、两栖类等, 两种方法所监测到的物种数量均远低于本研究。Li等(2012a)使用样线法仅监测到兽类16种, 表明该方法难以在固定线路监测到隐蔽性较强的动物, 在森林鸟类、小型兽类或夜行动物的监测上更为欠缺(Li et al, 2012b), 而且在实际调查过程中研究人员很少直接目击动物实体, 仅根据动物痕迹进行物种判断, 其准确性难以保证; 李成等(2017)使用围栏陷阱法在保护区内监测到21种脊椎动物, 但是其在实施过程中需要每天至少巡查一次陷阱, 以防天气原因导致的动物逃逸或死亡。如捕获到难以识别的物种, 还需将动物个体带回查证, 会对动物生活造成一定的影响(古晓东等, 2009)。在关于四川王朗国家级自然保护区物种监测方法的前期研究中, 还有铗夜法(刘少英等, 2001; 赵联军等, 2016)、遥感综合观测技术(李爱农等, 2018)等应用实例, 但是未见系统的数据发表, 本文未能做到一一对比。
综上, 没有一种单一的调查方法能完整监测到保护区内的所有动物物种, 每种方法都有其局限性, 受限于动物的体型、习性、生活史特征和栖息地条件(Li et al, 2012a)。在兽类、两栖类物种监测方面, DNA条形码法与红外线相机陷阱法、围栏陷阱法互为补充, 能最大限度地刻画比较全面的动物名录; 在鸟类物种方面, DNA条形码法可以补充红外相机难以拍摄到的小型林鸟的数据, 同时可以参考以往观鸟记录, 更直接地针对目标物种的适宜栖息地寻找其痕迹; 在爬行类物种方面, 尽管文献中记录铜蜓蜥(Sphenomorphus indicus)、菜花原矛头蝮(Protobothrops jerdonii)等在保护区有分布(刘少英等, 2001; 康东伟等, 2010), 但是本研究未采集到相关物种的样品, 有待探索更适合的监测方法(例如有针对性地设计样线、增加博物馆标本采样等)来丰富保护区该类群动物的条形码数据库。DNA条形码法主要依托于非损伤性取样, 相比于其他方法可以收集到更多样的动物样品, 准确率高, 且不对动物体本身造成损害, 在野生濒危动物的保护上具有重要的应用推广意义(魏辅文等, 2001; 俞可心等, 2022); DNA条形码法所产生的遗传数据, 结合形态学和生态学方法, 还会为评估物种多样性、明晰物种分布, 研究动物种群遗传结构、解析食物网和划定适宜的栖息地提供重要信息(Taberlet et al, 1999; Waits & Paetkau, 2005; Kress et al, 2015)。
附录 Supplementary Material
附录1 四川王朗国家级自然保护区DNA条形码资源库名录
Appendix 1 List of DNA barcoding bank in Wanglang National Nature Reserve, Sichuan
附录2 王朗齿突蟾DNA条形码序列
Appendix 2 DNA barcoding sequences of Scutiger Wanglangensis
致谢
感谢四川王朗国家级自然保护区傅晓波、罗春平、鲁超、梁春平、郑勇、李芯锐、赵和鑫、袁志伟等工作人员对野外调查工作的帮助, 感谢唐家河国家级自然保护区岷山山系动植物博物馆, 感谢四川大学生命科学学院/自然博物馆窦亮博士的鼎力支持。
参考文献
DNA detective: A review of molecular approaches to wildlife forensics
DOI:10.1007/s12024-009-9131-7
PMID:20013321
[本文引用: 1]

Illegal trade of wildlife is growing internationally and is worth more than USD$20 billion per year. DNA technologies are well suited to detect and provide evidence for cases of illicit wildlife trade yet many of the methods have not been verified for forensic applications and the diverse range of methods employed can be confusing for forensic practitioners. In this review, we describe the various genetic techniques used to provide evidence for wildlife cases and thereby exhibit the diversity of forensic questions that can be addressed using currently available genetic technologies. We emphasise that the genetic technologies to provide evidence for wildlife cases are already available, but that the research underpinning their use in forensics is lacking. Finally we advocate and encourage greater collaboration of forensic scientists with conservation geneticists to develop research programs for phylogenetic, phylogeography and population genetics studies to jointly benefit conservation and management of traded species and to provide a scientific basis for the development of forensic methods for the regulation and policing of wildlife trade.
四川王朗国家级自然保护区红喉雉鹑的日活动节律及种群密度
DNA commission of the International Society for Forensic Genetics: Guidelines for mitochondrial DNA typing
DNA barcoding meets molecular scatology: Short mtDNA sequences for standardized species assignment of carnivore noninvasive samples
DOI:10.1111/j.1755-0998.2011.03056.x
PMID:21883979
[本文引用: 1]

Although species assignment of scats is important to study carnivore biology, there is still no standardized assay for the identification of carnivores worldwide, which would allow large-scale routine assessments and reliable cross-comparison of results. Here, we evaluate the potential of two short mtDNA fragments [ATP6 (126 bp) and cytochrome oxidase I gene (COI) (187 bp)] to serve as standard markers for the Carnivora. Samples of 66 species were sequenced for one or both of these segments. Alignments were complemented with archival sequences and analysed with three approaches (tree-based, distance-based and character-based). Intraspecific genetic distances were generally lower than between-species distances, resulting in diagnosable clusters for 86% (ATP6) and 85% (COI) of the species. Notable exceptions were recently diverged species, most of which could still be identified using diagnostic characters and uniqueness of haplotypes or by reducing the geographic scope of the comparison. In silico analyses were also performed for a 110-bp cytochrome b (cytb) segment, whose identification success was lower (70%), possibly due to the smaller number of informative sites and/or the influence of misidentified sequences obtained from GenBank. Finally, we performed case studies with faecal samples, which supported the suitability of our two focal markers for poor-quality DNA and allowed an assessment of prey DNA co-amplification. No evidence of prey DNA contamination was found for ATP6, while some cases were observed for COI and subsequently eliminated by the design of more specific primers. Overall, our results indicate that these segments hold good potential as standard markers for accurate species-level identification in the Carnivora.© 2011 Blackwell Publishing Ltd.
Frequent mitochondrial gene introgression among high elevation Tibetan megophryid frogs revealed by conflicting gene genealogies
DOI:10.1111/j.1365-294X.2009.04258.x
PMID:19500253
[本文引用: 1]

Historical mitochondrial introgression causes differences between a species' mitochondrial gene genealogy and its nuclear gene genealogy, making tree-based species delineation ambiguous. Using sequence data from one mitochondrial gene (cytochrome b) and three nuclear genes (introns), we examined the evolutionary history of four high elevation Tibetan megophryid frog species, Scutiger boulengeri, Scutiger glandulatus, Scutiger mammatus and Scutiger tuberculatus. The three nuclear genes shared a similar history but the mitochondrial gene tree suggested a drastically different evolutionary scenario. The conflicts between them were explained by multiple episodes of mitochondrial introgression events via historical interspecific hybridization. 'Foreign' mitochondrial genomes might have been fixed in populations and extended through a large portion of the species' distribution. Some hybridization events were probably as old as 10 Myr, while others were recent. An F(1) hybrid was also identified. Historical hybridization events among the four species appeared to be persistent and were not restricted to the period of Pleistocene glaciation, as in several other well-studied cases. Furthermore, hybridization involved several species and occurred in multiple directions, and there was no indication of one mitochondrial genome being superior to others. In addition, incomplete lineage sorting resulting from budding speciation may have also explained some discrepancies between the mitochondrial DNA and nuclear gene trees. Combining all evidences, the former 'Scutiger mammatus' appeared to be two species, including a new species. With the availability of a wide range of highly variable nuclear gene markers, we recommend using a combination of mitochondrial gene and multiple nuclear genes to reveal a complete species history.
Impact of livestock terrain utilization patterns on wildlife: A case study of Wanglang National Nature Reserve
DOI:10.17520/biods.2019122
[本文引用: 1]

Terrain use is a fundamental factor of wildlife habitat and is closely related to other environmental factors. To assess the impact of cattle and horse on wildlife, we conducted field surveys in Wanglang National Nature Reserve and analyzed terrain preferences. We also deployed infrared cameras and historical biodiversity data to evaluate the risk of disturbance to key protected species based on altitudinal distribution of livestock. Results showed: (1) Both horse and cattle preferred low altitude habitats with low slopes, close to water with half to full sun aspects. However, there was also significant differences in terrain use between these two domestic animals. (2) The areas with the most frequent disturbance by livestock in the Wanglang Nature Reserve were Youyizhigou and Zhenggou in Zhugencha, and Yangdonggou in Dawodang. (3) Based on historical data, takin (Budorcas taxicolor tibetana) is likely the most influenced wildlife species, as the degree of overlap with livestock along altitudinal gradients is very high. Infrared camera data showed that areas frequented by livestock had fewer takin individuals recorded, indicating a negative relationship due to co-existence. Based on our findings, we suggest, (1) nature reserve should focus livestock in two core areas of Zhugencha (Youyizhigou, Zhenggou and Baishagou) and Dawodang (Yangdonggou and Waicepo) by monitoring the population and distribution of livestock in these areas; (2) they should prohibit the dispersal and distribution of livestock towards higher elevations; (3) the frequency with which local herdsman feed livestock salt needs to be controlled; (4) the strength of law enforcement towards illegal herds should be elevated.
基于地形的牲畜空间利用特征及干扰评价——以王朗国家级自然保护区为例
DOI:10.17520/biods.2019122
[本文引用: 1]

地形是栖息地的基本要素, 从地形评价动物的空间利用特征能够掌握动物的分布规律并进行预测。为掌握保护区内牲畜的空间利用特征, 并评价它们对主要保护动物的潜在影响, 我们于2018年5-11月调查了王朗国家级自然保护区内牛和马的分布, 并结合红外相机监测结果及历史监测数据进行了分析和评价。结果表明: (1)虽然两种牲畜均偏好低海拔、低坡度、光照良好(半阳坡、阳坡)、距水源近的栖息地, 但它们在地形利用上存在显著差异; (2)牲畜活动最频繁的三条沟分别是竹根岔右一支沟、竹根岔正沟和大窝凼洋洞沟, 且呈现不同的干扰特征; (3)基于监测数据, 羚牛(Budorcas taxicolor tibetana)可能是保护区内最易受牛马活动威胁的保护动物。红外相机监测结果显示, 羚牛沿海拔分布现状可能是回避牲畜密集区域的结果。基于本研究, 我们建议: (1)保护区重点关注竹根岔(右一支沟、正沟、白沙沟)、大窝凼(洋洞沟、外侧坡)两个核心区的牲畜活动情况, 并尽快针对放牧采取措施。例如, 持续监测重点干扰区域牲畜的种群数量和空间分布趋势。(2)严格限制牲畜继续向高海拔栖息地入侵。(3)管控放牧投盐等干扰的发生频率。(4)加强执法力度, 防止牲畜对保护区带来的干扰持续和扩大, 威胁物种安全。
Diversity, distribution and molecular species delimitation in frogs and toads from the Eastern Palaearctic
DOI:10.1093/zoolinnean/zlab083
URL
[本文引用: 2]

Biodiversity analyses can greatly benefit from coherent species delimitation schemes and up-to-date distribution data. In this article, we have made the daring attempt to delimit and map described and undescribed lineages of anuran amphibians in the Eastern Palaearctic (EP) region in its broad sense. Through a literature review, we have evaluated the species status considering reproductive isolation and genetic divergence, combined with an extensive occurrence dataset (nearly 85k localities). Altogether 274 native species from 46 genera and ten families were retrieved, plus eight additional species introduced from other realms. Independent hotspots of species richness were concentrated in southern Tibet (Medog County), the circum-Sichuan Basin region, Taiwan, the Korean Peninsula and the main Japanese islands. Phylogeographic breaks responsible for recent in situ speciation events were shared around the Sichuan Mountains, across Honshu and between the Ryukyu Island groups, but not across shallow water bodies like the Yellow Sea and the Taiwan Strait. Anuran compositions suggested to restrict the zoogeographical limits of the EP to East Asia. In a rapidly evolving field, our study provides a checkpoint to appreciate patterns of species diversity in the EP under a single, spatially explicit, species delimitation framework that integrates phylogeographic data in taxonomic research.
DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates
We describe "universal" DNA primers for polymerase chain reaction (PCR) amplification of a 710-bp fragment of the mitochondrial cytochrome c oxidase subunit I gene (COI) from 11 invertebrate phyla: Echinodermata, Mollusca, Annelida, Pogonophora, Arthropoda, Nemertinea, Echiura, Sipuncula, Platyhelminthes, Tardigrada, and Coelenterata, as well as the putative phylum Vestimentifera. Preliminary comparisons revealed that these COI primers generate informative sequences for phylogenetic analyses at the species and higher taxonomic levels.
A phylogeny of the high‐elevation Tibetan megophryid frogs and evidence for the multiple origins of reversed sexual size dimorphism
DOI:10.1111/jzo.2007.273.issue-3 URL [本文引用: 3]
Analysis and estimation for the landscapes of the nature as the Wanglang National Reserve
王朗自然保护区的大自然景观类型分析与评价
A simple and convenient method for measuring and monitoring small terrestrial vertebrates: Drift fence and pitfall traps
一种简便实用的小型陆栖脊椎动物监测方法——围栏陷阱法
Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator
Identification of birds through DNA barcodes
Molecular phylogenies indicate a Paleo-Tibetan origin of Himalayan lazy toads (Scutiger)
DOI:10.1038/s41598-017-03395-4
PMID:28607415
[本文引用: 1]

The Himalaya presents an outstanding geologically active orogen and biodiversity hotspot. However, our understanding of the historical biogeography of its fauna is far from comprehensive. Many taxa are commonly assumed to have originated from China-Indochina and dispersed westward along the Himalayan chain. Alternatively, the "Tibetan-origin hypothesis" suggests primary diversification of lineages in Paleo-Tibet, and secondary diversification along the slopes of the later uplifted Greater Himalaya. We test these hypotheses in high-mountain megophryid anurans (Scutiger). Extensive sampling from High Asia, and analyses of mitochondrial (2839 bp) and nuclear DNA (2208 bp), using Bayesian and Maximum likelihood phylogenetics, suggest that the Himalayan species form a distinct clade, possibly older than those from the eastern Himalaya-Tibet orogen. While immigration from China-Indochina cannot be excluded, our data may indicate that Himalayan Scutiger originated to the north of the Himalaya by colonization from Paleo-Tibet and then date back to the Oligocene. High intraspecific diversity of Scutiger implies limited migration across mountains and drainages along the Himalaya. While our study strengthens support for a "Tibetan-origin hypothesis", current sampling (10/22 species; 1 revalidated: S. occidentalis) remains insufficient to draw final conclusions on Scutiger but urges comparative phylogeographers to test alternative, geologically supported hypotheses for a true future understanding of Himalayan biogeography.
A new species of the toothed toad Oreolalax (Anura, Megophryidae) from Sichuan Province, China
DOI:10.3897/zookeys.929.49748
URL
[本文引用: 1]

The toad genus Oreolalax is widely distributed in southwest China and northern Vietnam. A new species of the genus is described from Sichuan Province, China. Phylogenetic analyses based on the mitochondrial 12S rRNA and 16S rRNA gene sequences supported the new species as an independent clade clustered into the clade also containing O. nanjiangensis and O. chuanbeiensis. The new species can be distinguished from its congeners by a combination of the following characters: body size moderate (SVL 51.2–64.2 mm in males); head broad; tympanum hidden; interorbital region with dark triangular pattern; belly with marbling; male lacking spines on lip margin; spiny patches on chest small with thick sparse spines in male; nuptial spines thick and sparse; tibio-tarsal articulation reaching beyond nostril when leg stretched forward; toe webbing at base.
History, current situation and prospects on nature reserves for giant pandas (Ailuropoda melanoleuca) in China
中国大熊猫保护区发展历史、现状及前瞻
大熊猫是我国的特产动物,古时被列为贡品,在近代被视为国宝。新中国成立以前,由于乱捕滥猎等因素,该物种已处于濒危状态。解放以后,我国政府于20 世纪50 年代开始了自然保护区建设,并于20 世纪60 年建立了卧龙等5 个大熊猫自然保护区,并将大熊猫列为禁猎动物。20 世纪70 年代我国进行了第一次全国大熊猫资源调查,基于调查结果新建了佛坪、唐家河、蜂桶寨等自然保护区,使有关大熊猫的自然保护区总数达到了13 个。1985 -1988 年全国第二次大熊猫资源调查结果显示,野生大熊猫种群数量较第一次减少了54 ﹪。进入90 年代,全国大熊猫保护区增至36 个,迄今共建立了63 个与保护大熊猫这一珍稀物种相关的自然保护区。全国大熊猫栖息地已由80 年代衰落期时22 220 km2 增加至34 000 km2,其中分布在保护区内的栖息地面积达29 000 km2,有效地保护了约85% 的大熊猫栖息地总面积以及约50% 的野生大熊猫种群数量。现存野生大熊猫种群数量估计约2 000只,因密度稀疏,发展空间大。目前,国家主管部门已出台了各种就地与迁地保护措施,只需各方通力合作,野生大熊猫的未来将是光明的。
An inexpensive, automation-friendly protocol for recovering high-quality DNA
DOI:10.1111/men.2006.6.issue-4 URL
China checklist of animals
In:
Analysis on management status of Wanglang Nature Reserve in Sichuan Province
四川王朗自然保护区管理状况分析
DNA barcodes for ecology, evolution, and conservation
DOI:10.1016/j.tree.2014.10.008 URL [本文引用: 3]
Conserve primers for sequencing complete ungulate mitochondrial cytochrome c oxidase I (COI) gene from problematic and decomposed biological samples
Space-air-field integrated biodiversity monitoring based on experimental station
DOI:10.17520/biods.2018052
[本文引用: 1]

Developing effective policies for biodiversity conservation and restoration policies requires spatially and temporally explicit data on distribution of species and habitats. Remote sensing provides an effective technical tool to meet this requirement. In recent years, the rapid development of integrated multi-platform, multi-scale, multi-mode remote sensing technology the implementation of integrated remote sensing observations across space-air-field provides novel opportunities for biodiversity monitoring. In this paper, we review the main methods of remote sensing that aids biodiversity monitoring and assess existing remote sensing observation experiments. We found that current methods of biodiversity monitoring using remotely-sensed data lacked the support of space-air-field integrated observations and the existing space-air-field integrated observations did not include biodiversity parameters. The Wanglang integrated observation and experiment station for mountain ecological remote sensing illustrates the potential to integrate experimental station-based and space-air-field integrated observations for biodiversity monitoring. Our review highlights that integrating direct observations with remote sensing can provide spatio-temporally explicit information on species and habitats and improve the informed monitoring of biodiversity.
基于站点的生物多样性星空地一体化遥感监测
DOI:10.17520/biods.2018052
[本文引用: 1]

科学制定生物多样性保护和恢复政策, 需要空间上连续、时间上高频的物种和生境分布以及物种迁移信息支持, 遥感是目前能满足该要求的有效技术手段。近年来, 遥感平台和载荷技术高速发展, 综合多平台、多尺度、多模式遥感技术, 开展基于站点的星空地一体化遥感观测试验, 可以对地表进行时空多维度、立体连续观测, 为生物多样性遥感监测提供了新的契机。本文总结了使用遥感技术监测生物多样性的主要方法, 回顾了典型的星空地一体化遥感观测试验。综述以往研究发现, 一方面, 现有遥感试验还缺少对生物多样性直接监测指标的观测, 另一方面, 生物多样性遥感监测方法也缺少星空地多维立体观测平台的支撑, 亟需加强两者的融合, 开展基于站点的生物多样性星空地一体化遥感监测研究。以设于我国四川王朗大熊猫国家级自然保护区内的王朗山地生态遥感综合观测试验站为例, 展示了星空地一体化遥感综合观测试验平台在生物多样性监测中的应用潜力。星空地一体化遥感观测可以提供物种和生境的综合定量信息, 与生态模型有机结合, 可以刻画生物多样性的时空格局与动态过程, 有助于挖掘过程机理, 提高生物多样性监测的信息化水平。
The relationship between community structure of ground- dwelling vertebrates and habitat types in the Wanglang Natural Reserve
王朗自然保护区地栖脊椎动物群落结构和生境类型的关系
Bird survey in Wanglang Nature Reserve
王朗自然保护区鸟类调查报告
A description of nests and Passeriformes bird eggs in Sichuan
四川雀形目鸟巢和鸟卵记述
Application of noninvasive sampling in conservation genetics
非损伤性取样法在保护遗传学研究中的应用
A direct comparison of camera-trapping and sign transects for monitoring wildlife in the Wanglang National Nature Reserve, China
DOI:10.1002/wsb.v36.3 URL [本文引用: 6]
Gauging the impact of management expertise on the distribution of large mammals across protected areas
DOI:10.1111/j.1472-4642.2012.00907.x URL [本文引用: 4]
The use of infrared-triggered cameras for surveying phasianids in Sichuan Province, China
DOI:10.1111/(ISSN)1474-919X URL [本文引用: 2]
Construction progress of the Camera-trapping Network for the Mountains of Southwest China
DOI:10.17520/biods.2020038 URL [本文引用: 1]
西南山地红外相机监测网络建设进展
Diversity of vertebrates in Wanglang Nature Reserve
王朗自然保护区脊椎动物多样性
Newly confirmed distribution of Volemys millicens in Wanglang
王朗自然保护区发现珍稀兽类四川田鼠
Improved COI barcoding primers for Southeast Asian perching birds (Aves: Passeriformes)
DOI:10.1111/j.1755-0998.2008.02221.x
PMID:21564563
[本文引用: 1]

The All Birds Barcoding Initiative aims to assemble a DNA barcode database for all bird species, but the 648-bp 'barcoding' region of cytochrome c oxidase subunit I (COI) can be difficult to amplify in Southeast Asian perching birds (Aves: Passeriformes). Using COI sequences from complete mitochondrial genomes, we designed a primer pair that more reliably amplifies and sequences the COI barcoding region of Southeast Asian passerine birds. The 655-bp region amplified with these primers overlaps the COI region amplified with other barcoding primer pairs, enabling direct comparison of sequences with previously published DNA barcodes.© 2008 The Authors. Journal compilation © 2008 Blackwell Publishing Ltd.
Diversity and distribution of culturable Mucoromycota fungi in the Greater Khinggan Mountains, China
DOI:10.17520/biods.2019058
[本文引用: 1]

To investigate the resources, diversity and distribution of culturable mucoromycotan fungi in the Greater Khingan Mountains, 279 samples including dry branches and fallen leaves, humus, soil and faeces were collected from nine representative counties and cities. A total of 1,153 strains of mucoromycotan fungi were isolated using a dilution plate, cutting and direct incubation methods. Among the 1,153 strains, 706 representatives were analyzed on the basis of fungal molecular barcode ITS rDNA. These mucoromycotan fungi were classified into 3 orders, 8 families, 10 genera and 38 species. Dominant genera were Mortierella, Umbelopsis, and Mucor, while dominant species were Mortierella amoeboidea, Mucor hiemalis, and Umbelopsis isabellina. This paper also summarized all 26 genera of Mucoromycotina and Mortierellomycotina reported in China, and the distribution of the dominant genera and species of the Greater Khinggan Mountains were analyzed. In addition, regional distribution of all genera was analyzed with regard to three major ecological regions (Eastern wet and semi-humid ecological regions, northwestern arid and semi-arid ecological regions and the Tibetan Plateau alpine ecological regions). The results showed that nine genera were distributed in all the three regions. For endemic genera, nine were found in the Eastern wet and semi-humid ecological region, only one in northwestern arid and semi-arid ecological region, and none in the Tibetan Plateau alpine ecological regions.
大兴安岭地区可培养毛霉门真菌多样性与分布
DOI:10.17520/biods.2019058
[本文引用: 1]

为查明大兴安岭地区可培养毛霉门真菌的资源、多样性及其分布, 本研究选取9个代表市县, 采集了279份枯枝落叶、腐殖质、土壤和粪便样品, 采用稀释平板挑取法、稀释平板切块法和样品直接培养挑取法进行分离培养。通过形态初步观察鉴定, 共得到毛霉门真菌1,153株。选取代表性的菌株706株, 基于真菌分子条形码ITS rDNA进行分子系统发育多样性分析, 明确了毛霉门真菌总计3目8科10属38种。优势属为被孢霉属(Mortierella)、伞形霉属(Umbelopsis)和毛霉属(Mucor), 优势种为类变形被孢霉(Mortierella amoeboidea)、冻土毛霉(Mucor hiemalis)和深黄伞形霉(Umbelopsis isabellina)。本文同时汇总了全国已报道毛霉亚门和被孢霉亚门共计26属的分布, 分析了大兴安岭优势属和优势种在全国的分布。按三大主要生态区(东部湿润、半湿润生态区, 西北干旱、半干旱生态区和青藏高原高寒生态区)对所有属进行区域分析, 结果表明: 有9个属在3个大区都有分布; 对特有属而言, 东部湿润、半湿润生态区发现9个, 西北干旱、半干旱生态区仅有1个, 青藏高原高寒生态区未发现。
A universal DNA mini-barcode for biodiversity analysis
DOI:10.1186/1471-2164-9-214
PMID:18474098
[本文引用: 1]

The goal of DNA barcoding is to develop a species-specific sequence library for all eukaryotes. A 650 bp fragment of the cytochrome c oxidase 1 (CO1) gene has been used successfully for species-level identification in several animal groups. It may be difficult in practice, however, to retrieve a 650 bp fragment from archival specimens, (because of DNA degradation) or from environmental samples (where universal primers are needed).We used a bioinformatics analysis using all CO1 barcode sequences from GenBank and calculated the probability of having species-specific barcodes for varied size fragments. This analysis established the potential of much smaller fragments, mini-barcodes, for identifying unknown specimens. We then developed a universal primer set for the amplification of mini-barcodes. We further successfully tested the utility of this primer set on a comprehensive set of taxa from all major eukaryotic groups as well as archival specimens.In this study we address the important issue of minimum amount of sequence information required for identifying species in DNA barcoding. We establish a novel approach based on a much shorter barcode sequence and demonstrate its effectiveness in archival specimens. This approach will significantly broaden the application of DNA barcoding in biodiversity studies.
Biodiversity hotspots for conservation priorities
DOI:10.1038/35002501 [本文引用: 1]
The Simple Fool's Guide to PCR, Version 2.0
From species to individuals: Combining barcoding and microsatellite analyses from non-invasive samples in plant ecology studies
DOI:10.1007/s11258-018-0866-7 [本文引用: 2]
Species identification by means of the cytochrome b gene
DOI:10.1007/s004140000134
PMID:11197623
[本文引用: 1]

Species identification was carried out by nucleotide sequence analysis of the cytochrome b (cytb) gene. The aim of the study was to identify biological specimens from diverse vertebrate animals by extracting and amplifying DNA from 44 different animal species covering the 5 major vertebrate groups (i.e. mammals, birds, reptiles, amphibians and fishes). The sequences derived were used to identify the biological origin of the samples by aligning to cytb gene sequence entries in nucleotide databases using the program BLAST. All sequences were submitted to the GenBank including new species which were not observed in the databases. The applicability of this method to the forensic field is demonstrated by simulated casework conditions where different types of samples including problematic specimens such as hair, bone samples, bristles and feathers were investigated to identify the species.
A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians
DOI:10.1016/j.ympev.2011.06.012
PMID:21723399
[本文引用: 1]

The extant amphibians are one of the most diverse radiations of terrestrial vertebrates (>6800 species). Despite much recent focus on their conservation, diversification, and systematics, no previous phylogeny for the group has contained more than 522 species. However, numerous studies with limited taxon sampling have generated large amounts of partially overlapping sequence data for many species. Here, we combine these data and produce a novel estimate of extant amphibian phylogeny, containing 2871 species (∼40% of the known extant species) from 432 genera (∼85% of the ∼500 currently recognized extant genera). Each sampled species contains up to 12,712 bp from 12 genes (three mitochondrial, nine nuclear), with an average of 2563 bp per species. This data set provides strong support for many groups recognized in previous studies, but it also suggests non-monophyly for several currently recognized families, particularly in hyloid frogs (e.g., Ceratophryidae, Cycloramphidae, Leptodactylidae, Strabomantidae). To correct these and other problems, we provide a revised classification of extant amphibians for taxa traditionally delimited at the family and subfamily levels. This new taxonomy includes several families not recognized in current classifications (e.g., Alsodidae, Batrachylidae, Rhinodermatidae, Odontophrynidae, Telmatobiidae), but which are strongly supported and important for avoiding non-monophyly of current families. Finally, this study provides further evidence that the supermatrix approach provides an effective strategy for inferring large-scale phylogenies using the combined results of previous studies, despite many taxa having extensive missing data.Copyright © 2011 Elsevier Inc. All rights reserved.
Phylogenetic relationships of the mustache toads inferred from mtDNA sequences
DOI:10.1016/j.ympev.2007.10.005 URL [本文引用: 1]
Identifying Rattus species using mitochondrial DNA
DOI:10.1111/men.2007.7.issue-5 URL [本文引用: 1]
DNA barcoding reveals 24 distinct lineages as cryptic bird species candidates in and around the Japanese Archipelago
DOI:10.1111/1755-0998.12282
PMID:24835119
[本文引用: 1]

DNA barcoding using a partial region (648 bp) of the cytochrome c oxidase I (COI) gene is a powerful tool for species identification and has revealed many cryptic species in various animal taxa. In birds, cryptic species are likely to occur in insular regions like the Japanese Archipelago due to the prevention of gene flow by sea barriers. Using COI sequences of 234 of the 251 Japanese-breeding bird species, we established a DNA barcoding library for species identification and estimated the number of cryptic species candidates. A total of 226 species (96.6%) had unique COI sequences with large genetic divergence among the closest species based on neighbour-joining clusters, genetic distance criterion and diagnostic substitutions. Eleven cryptic species candidates were detected, with distinct intraspecific deep genetic divergences, nine lineages of which were geographically separated by islands and straits within the Japanese Archipelago. To identify Japan-specific cryptic species from trans-Paleartic birds, we investigated the genetic structure of 142 shared species over an extended region covering Japan and Eurasia; 19 of these species formed two or more clades with high bootstrap values. Excluding six duplicated species from the total of 11 species within the Japanese Archipelago and 19 trans-Paleartic species, we identified 24 species that were cryptic species candidates within and surrounding the Japanese Archipelago. Repeated sea level changes during the glacial and interglacial periods may be responsible for the deep genetic divergences of Japanese birds in this insular region, which has led to inconsistencies in traditional taxonomies based on morphology.© 2014 John Wiley & Sons Ltd.
Noninvasive genetic analysis in birds: Testing reliability of feather samples
Opportunities and challenges of fecal DNA technology in molecular ecology researches
DOI:10.16829/j.slxb.150194
[本文引用: 1]

Advances in fecal DNA technology have expanded its usages in molecular ecology, especially in genetic assessment of wild animals. The technology allows researchers to understand ecological issues without contacting, disturbing, or even seeing animals, thus avoided invasions to the animal studied, and greatly promotes studies in molecular ecology of wild animals. Although this technology could yield poor DNA and relatively high genotyping errors in its early stages, these problems have been overcome gradually with the technological achievements being made in the past 25 years. Nowadays, fecal DNA technology yields good DNA and low genotyping errors, allowing researchers to address questions in reliability. Here, we share our knowledge about technological pitfalls on fecal sampling, preservation, DNA extraction, PCR, and genotyping in detail, and discuss opportunities and challenges of its applications, aiming to increase the power and role of the technology in molecular ecology of wild animals.<br>
粪便DNA分析技术在分子生态学研究中的机遇与挑战
Diversity and fauna composition of birds in the Wanglang National Nature Reserve, Sichuan
四川王朗国家级自然保护区鸟类多样性与区系组成
Generalist carnivores can be effective biodiversity samplers of terrestrial vertebrates
DOI:10.1002/fee.v19.10 URL [本文引用: 1]
Prey partitioning and livestock consumption in the world's richest large carnivore assemblage
DOI:10.1016/j.cub.2021.08.067 URL [本文引用: 1]
Fast surveys and molecular diet analysis of carnivores based on fecal DNA and metabarcoding
DOI:10.17520/biods.2018214
[本文引用: 5]

Large carnivores play an important role in the regulation of food-web structure and ecosystem functioning. However, large carnivores face serious threats that have caused declines in their populations and geographic ranges due to habitat loss and degradation, hunting, human disturbance and pathogen transmission. Conservation of large carnivore species richness and population size has become a pressing issue and an important research focus of conservation biology. The western Sichuan Plateau, located at the intersection of the mountains of southwest China and the eastern margin of the Tibetan Plateau, is a global biodiversity hotspot and has high carnivore species richness. However, increasing human activities may exacerbate the destruction of local flora and fauna, thereby threatening the survival of wild carnivores. Information on species composition and dietary habits can improve our understanding of the structure and function of the ecosystem and food-web relationships in the study area. In addition, species composition and dietary habits are of great significance for understanding multi-species coexistence mechanisms and preserving biodiversity. This study collected carnivore fecal samples from Xinlong and Shiqu counties in the Ganzi Tibetan Autonomous Prefecture, Sichuan Province. DNA was then extracted from the samples and the species was identified based on DNA sequences and DNA barcoding techniques. Seven carnivores were identified, including five large carnivores (Canis lupus, Ursus arctos, Panthera pardus, P. uncia and Canis lupus familiaris) and two medium and small-sized carnivores (Prionailurus bengalensis and Vulpes vulpes). Using fecal DNA, high-throughput sequencing and metabarcoding, we conducted diet analysis for the seven carnivores and found 28 different food molecular operational taxonomic units (MOTUs), including 19 mammals, eight birds and one fish species. The predominant prey categories of wolves, dogs and brown bears were ungulates. The domestic yak (Bos grunniens) was the most frequently identified prey species. Small mammals such as rodents and lagomorphs accounted for a significant proportion in the diets of leopard cats and red foxes, The most frequent prey of this category of carnivore were the Chinese scrub vole (Neodon irene) and plateau pika (Ochotona curzoniae). In addition, leopards and snow leopards mainly fed on the Chinese goral (Naemorhedus griseus) and blue sheep (Pseudois nayaur), respectively. Our study highlights the utility of fecal DNA and metabarcoding technique in fast carnivore surveys and high-throughput diet analysis, and provides a technical reference and guidance for future biodiversity surveys and food-web studies.
基于粪便DNA及宏条形码技术的食肉动物快速调查及食性分析
DOI:10.17520/biods.2018214
[本文引用: 5]

在陆地生态系统中, 大型食肉动物对于稳定食物网结构和生态系统功能有重要作用。在世界范围内, 由于栖息地丧失和破碎化、猎杀、人类活动干扰以及病原体的传播, 大型食肉动物生存正面临严重威胁, 多种食肉动物地理分布范围及种群数量大幅度缩减。如何有效保护大型食肉动物物种多样性及种群已经成为世界关注的焦点问题和保护生物学的重要研究方向。川西高原地处我国西南山地与青藏高原东缘交界地带, 属于世界生物多样性热点地区, 是世界大型食肉动物物种最丰富的地区之一, 而日益增强的人类活动可能会加剧对当地动植物资源的破坏, 进而威胁野生食肉动物的生存。获得准确的物种多样性信息及食肉动物食性数据有助于深入了解该地区生态系统结构及食物网关系, 对研究物种共存机制及生物多样性保护有重要意义。本研究通过从四川甘孜藏族自治州新龙县和石渠县野外采集的食肉动物粪便样品中提取DNA, 利用DNA条形码进行物种鉴定, 快速获得该地区食肉动物物种构成信息。38份粪便样品经鉴定来自于7种食肉动物, 分别为5种大型食肉动物(狼Canis lupus、棕熊Ursus arctos、豹Panthera pardus、雪豹P. unica、狗Canis lupus familiaris)和2种中小型食肉动物(豹猫Prionailurus bengalensis、赤狐Vulpes vulpes)。进一步利用高通量测序和宏条形码技术对7种食肉动物粪便中的食物DNA进行精准食性分析, 得到包含19种哺乳类、8种鸟类和1种鱼类共计28个不同的食物分子可操作分类单元(molecular operational taxonomic unit, MOTU)。结果显示, 狼、狗、棕熊最主要的食物来源为偶蹄目动物, 其中取食频率最高的物种为家牦牛(Bos grunniens); 而豹猫和赤狐食物中小型哺乳动物如啮齿目和兔形目占重要比例, 其中高原松田鼠(Neodon irene)和高原鼠兔(Ochotona curzoniae)被取食频率最高。豹和雪豹的食物分别为偶蹄目的中华斑羚(Naemorhedus griseus)和岩羊(Pseudois nayaur)。本研究显示了粪便DNA及宏条形码技术在食肉动物多样性快速调查及高通量精确食性分析中的应用前景, 并为此类研究提供了技术路线的有力借鉴。
Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers
DOI:10.1093/aesa/87.6.651 URL [本文引用: 1]
Relative efficiencies of the maximum parsimony and distance-matrix methods in obtaining the correct phylogenetic tree
The relative efficiencies of the maximum parsimony (MP) and distance-matrix methods in obtaining the correct tree (topology) were studied by using computer simulation. The distance-matrix methods examined are the neighbor-joining, distance-Wagner, Tateno et al. modified Farris, Faith, and Li methods. In the computer simulation, six or eight DNA sequences were assumed to evolve following a given model tree, and the evolutionary changes of the sequences were followed. Both constant and varying rates of nucleotide substitution were considered. From the sequences thus obtained, phylogenetic trees were constructed using the six tree-making methods and compared with the model (true) tree. This process was repeated 300 times for each different set of parameters. The results obtained indicate that when the number of nucleotide substitutions per site is small and a relatively small number of nucleotides are used, the probability of obtaining the correct topology (P1) is generally lower in the MP method than in the distance-matrix methods. The P1 value for the MP method increases with increasing number of nucleotides but is still generally lower than the value for the NJ or DW method. Essentially the same conclusion was obtained whether or not the rate of nucleotide substitution was constant or whether or not a transition bias in nucleotide substitution existed. The relatively poor performance of the MP method for these cases is due to the fact that information from singular sites is not used in this method. The MP method also showed a relatively low P1 value when the model of varying rate of nucleotide substitution was used and the number of substitutions per site was large. However, the MP method often produced cases in which the correct tree was one of several equally parsimonious trees. When these cases were included in the class of "success," the MP method performed better than the other methods, provided that the number of nucleotide substitutions per site was small.
Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding
Noninvasive genetic sampling: Look before you leap
DOI:10.1016/S0169-5347(99)01637-7 URL [本文引用: 2]
MEGA11: Molecular evolutionary genetics analysis version 11
DOI:10.1093/molbev/msab120
PMID:33892491
[本文引用: 1]

The Molecular Evolutionary Genetics Analysis (MEGA) software has matured to contain a large collection of methods and tools of computational molecular evolution. Here, we describe new additions that make MEGA a more comprehensive tool for building timetrees of species, pathogens, and gene families using rapid relaxed-clock methods. Methods for estimating divergence times and confidence intervals are implemented to use probability densities for calibration constraints for node-dating and sequence sampling dates for tip-dating analyses, which will be supported by new options for tagging sequences with spatiotemporal sampling information, an expanded interactive Node Calibrations Editor, and an extended Tree Explorer to display timetrees. We have now added a Bayesian method for estimating neutral evolutionary probabilities of alleles in a species using multispecies sequence alignments and a machine learning method to test for the autocorrelation of evolutionary rates in phylogenies. The computer memory requirements for the maximum likelihood analysis are reduced significantly through reprogramming, and the graphical user interface (GUI) has been made more responsive and interactive for very big datasets. These enhancements will improve the user experience, quality of results, and the pace of biological discovery. Natively compiled GUI and command-line versions of MEGA11 are available for Microsoft Windows, Linux, and macOS from www.megasoftware.net.© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Preliminary surveys of wild animals using infrared camera in Wanglang National Nature Reserve, Sichuan Province
DOI:10.17520/biods.2017082
[本文引用: 5]

We used infrared camera to monitor wild animals in Wanglang National Nature Reserve. Our goal was to estimate species diversity, the relationship between the number of cameras and number of species, the relationship between the camera days and number of species, and the relative abundance of species. Across 42 locations, we recorded a total of 1,793 images in which we found 25 species of wildlife. Species found in our camera traps included three national first-class protected species such as giant panda (Ailuropoda melanoleuca), Sichuan takin (Budorcas tibetanus) and buff-throated partridge (Tetraophasis szechenyii). We also recorded eight national second-class protected wild animals such as the Asiatic black bear (Ursus thibetanus), yellow-throated marten (Martes flavigula), Chinese serow (Capricornis milneedwardsii), Chinese goral (Naemorhedus griseus). These 25 species were captured when we increased the number of trap-days increased to 23 or any single the trap was placed for at least 180 days. Furthermore the blood pheasant (Ithaginis cruentus) (29.28) and the tufted deer (Elaphodus cephalophus) (15.78) had the highest relative abundances in the reserve. The relative abundance index for the giant panda was 8.09 and the indices for temminck’s tragopan (Tragopan temminckii), Chinese serow and buff-throated partridge was between 2 and 5. The Chinese goral, koklass pheasant (Pucrasia macrolopha), Asiatic black bear, Sichuan takin and blue eared-pheasant (Crossoptilon auritum) had the lowest relative abundance values (all < 1). In summary, we found camera-trapping to be an effective method for wildlife survey. Our findings suggest that less abundant species need more camera trapping effort. Nature reserves are important for protecting habitats of endangered species.
野生动物物种多样性的初步调查
DOI:10.17520/biods.2017082
[本文引用: 5]

本研究以四川王朗国家级自然保护区为研究区域, 利用红外相机对保护区内的主要野生动物进行了初步调查, 分析了该区域的物种多样性现状、相机数量和相机工作日与物种数量间的关系以及物种的相对丰富度。结果表明: 42台红外相机共拍摄到物种独立照片1,793张, 鉴定出野生动物25种, 包括大熊猫(Ailuropoda melanoleuca)、四川羚牛(Budorcas tibetanus)和黄喉雉鹑(Tetraophasis szechenyii) 3种国家一级重点保护野生动物, 黑熊(Ursus thibetanus)、黄喉貂(Martes flavigula)、中华鬣羚(Capricornis milneedwardsii)、中华斑羚(Naemorhedus griseus)等8种国家二级重点保护野生动物。在相机数量增加到23台的时候拍摄到了本次记录的全部25种野生动物, 并且在单台相机工作日达到180天时, 物种数达到饱和。保护区内物种相对丰富度最高的是血雉(Ithaginis cruentus)(29.28)和毛冠鹿(Elaphodus cephalophus)(15.78); 大熊猫的相对丰富度为8.09; 红腹角雉(Tragopan temminckii)、中华鬣羚和黄喉雉鹑的相对丰富度在2-5之间; 中华斑羚、勺鸡(Pucrasia macrolopha)、黑熊、四川羚牛和蓝马鸡(Crossoptilon auritum)的相对丰富度最低, 不到1。综上所述, 红外相机能够有效地对野生动物资源进行监测调查, 对于相对丰富度较低的物种需要投入更多的精力, 这些物种的栖息地保护对于自然保护区的发展至关重要。
Impacts of livestock grazing, topography and vegetation on distribution of wildlife in Wanglang National Nature Reserve, China
The diversity of large- and medium-sized terrestrial mammals and birds in the Giant Panda National Park: A meta-analysis based on camera-trapping data
DOI:10.17520/biods.2021165
[本文引用: 1]

<p id="p00015"><strong>Aims</strong> Biodiversity monitoring is the foundation of conservation work in national parks. Systematic conservation planning and effective management actions within these parks highly rely on an in-depth evaluation of biodiversity metrics. In China, the Giant Panda National Park (GPNP) is one of the first five national parks. To establish baseline metrics of mammal and bird diversity in GPNP, we conducted a meta-analysis based on published camera-trapping data. <br><strong> Methods</strong> We systematically searched academic publications, project reports, and news articles that reported on wildlife camera-trapping studies between 2005 and 2020 in GPNP. We also conducted a questionnaire survey on the history and results of camera-trap monitoring projects within the protected areas of the region. These data were compiled for statistical analysis. <br><strong> Result</strong> Between 2005 and 2020, 71 wild mammal species belonging to 6 orders, 22 families and 55 genera, and 232 wild bird species belonging to 13 orders, 45 families and 132 genera were recorded via camera-trap monitoring in 51 protected areas within GPNP. Among the four mountain ranges within GPNP (i.e., Mts. Qinling, Mts. Minshan, Mts. Qionglai and Mts. Xiangling), the species richness of large- and medium-sized terrestrial mammals and birds was the highest in Mts. Qionglai and Mts. Minshan (40 mammal and 12 pheasant species for each) and the lowest in Mts. Xiangling (25 mammal and 7 pheasant species). The recorded number of target species in individual protected areas was positively correlated with the area of protected area, sampling effort (measured as number of camera-days), and the camera station elevation range. The numbers of species recorded in national protected areas (28 ± 8.3, mean ± SD) were significantly higher than those in provincial protected areas (19 ± 8.9). Four large carnivores of Felidae and Canidae (leopard <i>Panthera pardus</i>, snow leopard <i>P. uncia</i>, wolf <i>Canis lupus</i> and dhole <i>Cuon alpinus</i>) were recorded in GPNP, primarily from Mts. Qinling and Mts. Qionglai, while no large carnivores were recorded within the park in Mts. Minshan and only one wolf was recorded in Mts. Xiangling. <br><strong> Conclusion</strong> The previous protected area network and camera-trapping monitoring network prior to the establishment of GPNP have already accumulated a high quantity of data on wild mammals and birds in this region. These data provide a reliable baseline biodiversity inventory for the pilot and construction phases of GPNP. In light of these results, GPNP should design and implement a standardized wildlife monitoring system to further provide additional data for future evaluations of park management, decision-making, and conservation effectiveness.</p>
大熊猫国家公园的地栖大中型鸟兽多样性现状: 基于红外相机数据的分析
DOI:10.17520/biods.2021165
[本文引用: 1]

生物多样性监测是国家公园保护的核心基础。大熊猫国家公园是我国首批5个国家公园之一, 系统的保护规划与有效的管理行动均有赖于对区内生物多样性本底、现状与动态的深入了解。为了解大熊猫国家公园范围内兽类与鸟类多样性本底与现状, 本研究系统检索了该区域内2005-2020年基于红外相机调查技术的野生动物研究论文、项目报告以及新闻报道, 并对区内原有保护地的红外相机监测历史与结果进行了问卷调查。结果表明, 2005-2020年期间, 在大熊猫国家公园范围内51个保护地的红外相机调查与监测中, 共记录到分属6目22科55属的71种野生兽类与分属13目45科132属的232种野生鸟类。在国家公园所覆盖的秦岭、岷山、邛崃山、相岭4大山系中, 邛崃和岷山记录到的大中型地栖鸟兽物种多样性最高(均为兽类40种, 鸟类12种), 相岭最低(兽类25种, 鸟类7种)。单个保护地中记录到的大中型地栖鸟兽物种数量与保护地面积、红外相机有效工作日及相机位点的海拔跨度均呈正相关, 国家级保护地中记录到的物种数(28 ± 8.3, mean ± SD)显著高于省级保护地(19 ± 8.9)。在大熊猫国家公园内共记录到猫科与犬科的4种大型食肉动物, 即豹(Panthera pardus)、雪豹(P. uncia)、狼(Canis lupus)和豺(Cuon alpinus), 主要来自于秦岭山系和邛崃山系, 而国家公园内的岷山山系则没有记录到大型食肉动物, 相岭山系中仅有1次狼的记录。本研究结果显示, 大熊猫国家公园内前期已经建立起的自然保护地网络与红外相机监测体系已积累大量区内野生兽类与鸟类的基础数据, 为国家公园的试点与建设提供了生物多样性编目与监测方面的可靠本底。在这些前期工作的基础上, 大熊猫国家公园应进一步规划、建设标准化的野生动物监测体系, 为今后国家公园的管理决策、成效评估提供坚实的科学支撑。
DNA barcoding for ecologists
DOI:10.1016/j.tree.2008.09.011 URL [本文引用: 3]
Noninvasive genetic sampling tools for wildlife biologists: A review of applications and recommendations for accurate data collection
DOI:10.2193/0022-541X(2005)69[1419:NGSTFW]2.0.CO;2 URL [本文引用: 2]
Molecular scatology and its application—Reliability, limitation and prospect
分子粪便学及其应用——可靠性、局限性和展望
Supplement of small mammals in Wanglang Nature Reserve of Pingwu County
平武县王朗自然保护区小型兽补遗
A list of fleas and mites in vitro from small mammals in Wanglang Nature Reserve of Pingwu, Sichuan Province
四川省平武王朗自然保护区小型兽体外蚤、螨名录
Progress and prospect of China biodiversity monitoring from a global perspective
DOI:10.17520/biods.2022434
[本文引用: 1]

Background & Aim: Analyzing biodiversity status requires multi-spatial scale, continuous monitoring across different ecosystems due to its heterogenous nature in both space and time. Therefore, monitoring networks are necessary for biodiversity conservation research. Biodiversity monitoring networks at the global, regional, and national scales, represented by GEO BON and APBON, have flourished. China has established a long-term monitoring network for ecosystems and species at the national scale. and the China Biodiversity Observation and Research Network (Sino BON) was launched in 2013 with strong support from the Chinese Academy of Sciences and the Ministry of Finance. Review Results: Sino BON includes 10 subnetworks specialized at monitoring animals, plants and microbes and an additional network for near-ground remote sensing, which covers 30 main sites and 60 affiliated sites in China. Currently, Sino BON has created a research platform for multi-trophic interactions among soil microorganisms, insects, large mammals, underground forests to forest canopies. This platform provides an understanding of biodiversity change and its driving factors at the national level and may be used in protecting biodiversity and sustainable utilization of biological resources. Perspectives: For further progresses, monitoring technology, monitoring areas, data standards and integrated information platforms require further development.
全球视角下的中国生物多样性监测进展与展望
DOI:10.17520/biods.2022434
[本文引用: 1]

生物多样性强烈的时空尺度依赖性和多层次性决定了生物多样性现状与变量的分析需要在不同生态系统进行多空间尺度、全面和连续的监测。因此, 构建生物多样性研究监测网络是生物多样性保护和研究的基础工作。近年来, 对地观测组织-生物多样性观测网络(GEO BON)、亚太生物多样性监测网络(APBON)等全球、区域以及国家尺度的生物多样性监测网络蓬勃发展。中国陆续在国家尺度上建立了针对生态系统和物种的长期监测网络, 其中, 中国生物多样性监测与研究网络(China Biodiversity Observation and Research Network, Sino BON)于2013年启动建设, 在我国主要生态系统和环境梯度设置30个监测主点和60个监测辅点, 目前已建成10个专项网对动物、植物和微生物进行监测, 并建立了以数据标准与汇交、近地面遥感为核心的综合监测中心。Sino BON打造了从地下、地面到森林林冠的多尺度、多类群(功能群)以及多营养级交互为重点的监测与研究平台, 为理解生物多样性变化趋势及其驱动因素、研究生物多样性维持机制, 以及国家履行《生物多样性公约》、保护生物多样性和生物资源提供详实可靠的生物多样性变化数据。为进一步支撑国家生物多样性治理能力、深化全球多样性保护合作, 我国生物多样性监测亟需在监测技术、监测区域、数据标准、综合信息平台等方向谋求更大的发展。
Molecular analysis of vertebrates and plants in scats of leopard cats (Prionailurus bengalensis) in southwest China
DOI:10.1093/jmammal/gyw061
URL
[本文引用: 1]

Understanding the diets of carnivores is essential for resolving food web interactions and the population dynamics of both prey and predators and for designing effective strategies for species and ecosystem conservation. As effective predators, wild felids play important roles in various ecosystems, but relatively little is known about the dietary habits of many species, primarily owing to their elusive behavior. We used a DNA-based method to analyze the vertebrates and plants constituting the diet of leopard cats (Prionailurus bengalensis) in the temperate forests of the mountains of southwest China, a global biodiversity hotspot. DNA extracted from leopard cat scats was amplified with primers targeting either the vertebrate mitochondrial 12S rRNA gene (N = 25 scats) or the psbCL region of the chloroplast genome of vascular plants (N = 42 scats). The polymerase chain reaction products were sequenced and prey taxa were assigned based on sequence similarity. We identified a total of 16 taxa of vertebrate prey, with pikas (in 76% of the scats) and rodents (40%) predominating. Plant material belonging to 12 taxa was found in 76% of the samples, and the genus Solanum and subfamily Rosoideae were the most frequently detected plant taxa. The frequency of occurrence of identified plant taxa differed between the spring–summer and fall–winter months. Thus, the leopard cats in our study area have a diversified diet and are highly flexible in adapting their foraging behavior to the locally available prey. Our data suggest that preserving their natural prey base dominated by pikas and rodents may be vital for the subsistence of local populations. The high species resolution and detection sensitivity of the DNA-based method we used in this study make it a powerful and efficient tool for fine-scale analysis of complex diets.
A new species of Megophryidae—Scutiger (Scutiger) wanglangensis from Sichuan, China (Amphibia, Anura)
四川角蟾科(Megophryidae)一新种——王朗齿突蟾(两栖纲: 无尾目)
Research progress on wildlife identification technology
野生动物鉴定技术研究进展
Using Noninvasive Genetic Sampling to Estimate the Population Size and Study Dispersal of Giant Pandas
利用非损伤性遗传取样研究大熊猫的种群数量和扩散模式
A simple protocol for DNA extraction from faeces of the giant panda and lesser panda
大熊猫和小熊猫粪便DNA提取的简易方法
A survey of small mammals in Wanglang Nature Reserve
王朗自然保护区小型兽类的调查
Daytime driving decreases amphibian roadkill
Genetic diversity and conservation of endangered animal species
DOI:10.1351/pac200274040575
URL
[本文引用: 1]

The loss of biodiversity resulting from extinctions is receiving increasing attention. Over several thousands of animal species have been evaluated and recognized as endangered species. Inbreeding depression has been demonstrated in many wild animal species. Here we sequenced 655-978 bp mitochodrial D-loop region of 32 individuals from four regional giant panda populations. Sixteen haplotypes were observed. AMOVE analysis demonstrated that genetic differentiation was not significant in the overall population, except the Qingling population. The current panda population may recover from a recent severe bottleneck that occurred about 43 000 years ago. Combining with our results on two endangered snub-nosed monkey species and one common hare species, different scenarios for low genetic variation have been discussed. Our results suggest that low genetic variation does not necessary result from a recent bottleneck, and it is not necessarily an indication of the level of endangerment.
A study of diversities of small mammals in Wanglang National Nature Reserve
王朗国家级自然保护区小型兽类多样性研究
Phylogenetic relationships of megophryid genera (Anura: Megophryidae) based on partial sequences of mitochondrial 16S rRNA gene
基于16S rRNA序列的角蟾科部分属间系统关系