然而, 珍稀濒危动物较低的种群密度及难以预测的行为活动使得研究者很难通过直接观察获得食性信息。传统的食性研究方法主要以目标物种的粪便为实验材料, 对粪便中未消化的食物残渣进行形态学分析以获取食物组成信息(Klare et al, 2011; Monterroso et al, 2019)。尽管这种研究方法具有低成本、易操作以及非损伤性采样等优点, 但存在较多的局限性(Klare et al, 2011)。如需要分析大量的粪便样品且十分依赖于粪便中食物残渣的可辨识度(Trites & Joy, 2005), 当食物形态相近、被彻底消化或无硬质残留时, 就会造成结果的偏差(Carss & Parkinson, 1996)。同时, 该方法对研究人员根据形态学特征进行物种鉴定的能力要求很高, 受主观因素影响较大, 过程繁杂且准确性和可重复性也难以保证(Spaulding et al, 2000)。
得益于分子生物学技术的发展, 高通量测序(high-throughput sequencing, HTS)和DNA宏条形码技术(DNA metabarcoding)的出现为食性研究提供了一种更高效、准确的方法(Pompanon et al, 2012; Taberlet et al, 2012; Monterroso et al, 2019)。DNA宏条形码技术是利用高通量测序技术大量获得样本中多种条形码基因序列, 继而通过生物信息学手段实现分类单元鉴定的方法(邵昕宁等, 2019)。它对粪便中稀少、高度降解、形态相近、无硬质部分的食物残渣具有高度敏感性, 在获取更大尺度的物种信息的同时避免了传统分类学的人工误差(Monterroso et al, 2019; Traugott et al, 2020)。目前, 该技术已成功应用于欧亚水獭(Lutra lutra)及其他食肉目动物的食性研究中(Xiong et al, 2017; Kumari et al, 2019; Marcolin et al, 2020; Wang et al, 2022)。
作为淡水生态系统的顶级捕食者之一, 欧亚水獭因其对水质的高度敏感性, 被普遍作为淡水生态系统的指示种和旗舰种(Ruiz et al, 1998; Kruuk, 2006)。自20世纪中期以来, 受盗猎、栖息地破坏、环境污染等人为干扰影响, 我国欧亚水獭种群已遭受严重破坏, 部分地区甚至已野外灭绝(徐龙辉, 1984; Gomez et al, 2016; Li & Chan, 2018)。为保护其种群, 在1988年欧亚水獭就已被《国家重点保护野生动物名录》列为国家二级重点保护野生动物。有研究表明, 欧亚水獭现主要分布于我国东北和西南地区, 其中大兴安岭是维持东北地区水獭种群稳定的关键区域(吕江等, 2018; Zhang et al, 2018; 张超等, 2022)。目前国内欧亚水獭的研究主要集中在种群密度(吕江等, 2018; Zhang et al, 2018; 张超等, 2022)、活动节律(韩雪松等, 2021; 史国强等, 2021)、生境利用(Wang et al, 2021)等领域。然而我国水獭的食性研究仍处于起步阶段, 仅在四川唐家河自然保护区开展过基于粪便的水獭食性研究(Wang et al, 2022), 东北地区欧亚水獭的食性研究尚且空白。
本研究以大兴安岭北部欧亚水獭指名亚种(Lutra lutra lutra)野生种群的粪便为实验材料, 利用DNA宏条形码技术研究其食物组成以完善我国东北地区欧亚水獭野生种群食性的基础信息, 为评估其生存状况以及制定保护措施提供科学依据。
1 材料与方法
1.1 研究区域
1.2 样品采集和保存
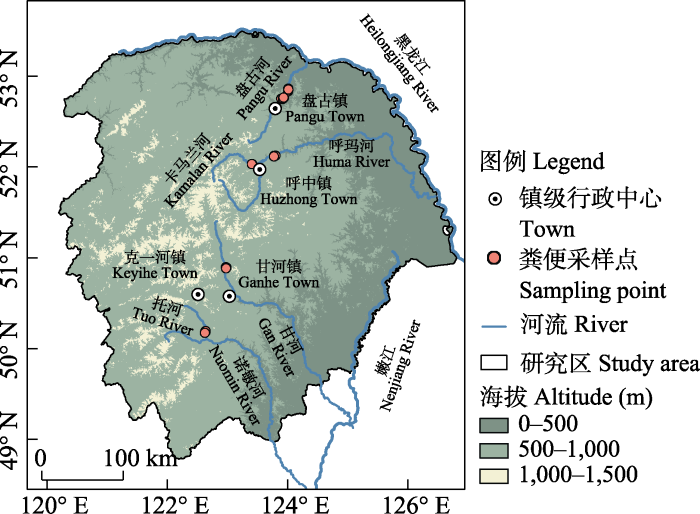
图1
图1
大兴安岭北部欧亚水獭粪便样品采集地点
Fig. 1
The locations of Eurasian otter feces collected in the northern Greater Khingan Mountains
PCR扩增体系为25 μL, 包括样品DNA 30 ng、2×Taq Plus Master Mix 12.5 μL、正反引物各1 μL (5 μM)以及2 ng/μL牛血清蛋白(BSA) 3 μL, ddH2O补充至25 μL。PCR反应程序为94℃预变性5 min; 94℃变性30 s, 50℃退火30 s, 72℃延伸60 s, 循环35次; 72℃延伸7 min。
采样时工作人员沿样线进行调查, 根据欧亚水獭活动痕迹及粪便特征辨认目标样品。采集样品时, 佩戴一次性医用乳胶手套, 每次拾取样品后更换新的手套以避免交叉污染, 记录GPS坐标、采样时间以及相关环境信息。每份样品分别装入已加入8倍于粪便样品体积无水硅胶的50 mL离心管中干燥保存(Wasser et al, 1997), 带回实验室后转移至-20℃低温保存。
1.3 粪便DNA提取、扩增及物种鉴定
粪便样品使用QIAamp Fast DNA Stool Mini Kit DNA提取试剂盒提取DNA, 然后使用Nano Drop 2000 (Thermo Scientific)测定DNA浓度和纯度, 选用合格样品进行物种鉴定。
为了确定每份粪便样品的来源为欧亚水獭, 利用欧亚水獭特异引物LLCBL/R (表1)对粪便中线粒体DNA Cytb基因片段进行扩增, 扩增产物长度为80 bp。
表1 本研究使用的引物序列
Table 1
| 引物 Primer | 扩增区域 Target | 引物序列 Primer sequence (5'-3') | 产物长度 Product length | 参考文献 Reference |
|---|---|---|---|---|
| LLCBL/R | Cytb | F: AAAGCCACCCTGACACGATT | 80 bp | Thomsen et al, 2012 |
| R: AGCAGGTGGATTGTTGCTAGTG | ||||
| 12S V5 | 12S rRNA | F: TAGAACAGGCTCCTCTAG | 98 bp | Riaz et al, 2011 |
| R: TTAGATACCCCACTATGC | ||||
| OBS1 | - | CTATGCTCAGCCCTAAACATAGATAGCTTACATAACAAAACTATCTGC-C3 | - | Kumari et al, 2019 |
| HomoB | - | CTATGCTTAGCCCTAAACCTCAACAGTTAAATCAACAAAACTGCT-C3 | - | De Barba et al, 2014 |
| BF2/BR1 | COI | F: GCHCCHGAYATRGCHTTYCC | 322 bp | Elbrecht & Leese, 2017 |
| R: ARYATDGTRATDGCHCCDGC |
PCR产物使用1%琼脂糖凝胶电泳进行检测, 以欧亚水獭肌肉样品为阳性对照, 以粪便DNA提取空白为阴性对照, 筛选出目标条带清晰、长度为80 bp的样品进行食性分析。
1.4 食性分析的PCR扩增及高通量测序
1.4.1 食性分析的PCR扩增
确定粪便来源物种后, 进行基于DNA宏条形码的分子食性分析。选用针对脊椎动物设计的通用引物12S V5-F/12S V5-R (表1)对粪便中脊椎动物线粒体基因12S rRNA V5片段进行扩增, 扩增片段长度为98 bp (Riaz et al, 2011)。该对引物通用性广且具有较高的种属分辨率, 已成功应用于欧亚水獭、雪豹(Panthera uncia)、豹猫(Prionailurus bengalensis)等多种食肉动物的分子食性分析中(Xiong et al, 2017; Kumari et al, 2019; Harper et al, 2020)。使用该组引物进行PCR扩增时, 能够同时扩增出粪便中欧亚水獭及其食物的DNA, 为减少欧亚水獭DNA以及潜在人类DNA竞争性扩增对食物DNA扩增的影响, 本研究使用欧亚水獭阻隔引物(Eurasian otter blocking) OBS1以及人类阻隔引物(human blocking) HomoB来抑制欧亚水獭与人类DNA的扩增(Kumari et al, 2019)。
PCR扩增体系为25 μL, 包括样品DNA 30 ng、2×Taq Plus Master Mix 12.5 μL、正反引物各1 μL (5 μM) 以及2 ng/μL牛血清蛋白(BSA) 3 μL, 另外在12S V5引物体系中加入阻隔引物(50 μM) 1 μL, 最后ddH2O补充至25 μL。PCR反应程序为94℃预变性5 min; 94℃变性30 s, 12S V5引物体系60℃退火30 s, BF2/BR1引物体系50℃退火30 s, 72℃延伸60 s, 进行35个循环; 72℃延伸7 min。同时在每批次扩增时加入粪便DNA提取空白和不加模板DNA的PCR空白作为对照以检验DNA污染情况。
1.4.2 高通量测序及数据处理
PCR产物使用1%琼脂糖凝胶电泳检测扩增条带大小, 并用Agencourt AMPure XP核酸纯化试剂盒进行纯化, 然后构建DNA测序文库进行高通量测序。测序采用双端测序法(paired-end sequencing), 每端读长300 bp, 使用平台为Illumina Miseq PE300系统(Illumina Inc., San Diego, CA, USA), 由北京奥维森基因科技有限公司完成。
测序完成后, 使用Trimmomatic (v0.36)、Pear (v0.9.6)对原始数据进行质量控制并去除含N碱基的DNA序列。之后在QIIME 2软件(2021.11.0) (Bolyen et al, 2019)中使用vsearch (Rognes et al, 2016)根据双端序列的overlap关系进行拼接处理。拼接后, 使用DADA2 (Callahan et al, 2016)对合并序列进行质量控制、降噪、去嵌合体, 然后对序列进行聚类形成扩增序列变体(amplicon sequence variant, ASV, 类似以100%相似度聚类的可操作分类单元(operational taxonomic units, OTU))。
基于BLAST局域比对的序列物种鉴定及分类原则包括: (1)比对结果一致度 ≥ 98%时, 最匹配序列只对应单一物种, 且该物种在当地有分布, 则认为序列来自该物种, 当该物种为非当地物种时, 则记为涵盖当地最近缘物种的上一级分类单元; 最匹配序列对应多物种时, 先排除当地无分布的物种, 如仍对应不止一种, 则注释结果记为涵盖这些物种的最小分类单元, 若物种均非当地物种时, 则记为涵盖当地最近缘物种的最小分类单元。(2)当95% ≤ 比对结果一致度 < 98%时, 最匹配序列仅有1条, 且与其次之的序列一致度差距2%时, 则记为一致度最高序列的上一级分类单元; 最匹配序列对应多个物种时, 则记为能涵盖当地最近缘物种的最小分类单元。(3)比对结果一致度 < 95%时, 序列物种注释结果记为未知。
对使用针对无脊椎动物的BF2/BR1引物组扩增的序列数据, 使用基于朴素贝叶斯原理的RDP物种分类方法进行注释, 参考数据库从blast比对结果中构建(Schmiedova et al, 2022)。具体方法为: 使用BLAST+ (2.13.0)将实验中获得的ASV与nt数据库进行比对, 提取前250条以一致度由高到低排序的同源序列组成参考数据库。使用QIIME2软件中的feature-classifier classify-sklearn (Bokulich et al, 2018), 对ASV使用基于朴素贝叶斯原理的RDP物种分类器进行分类, 分类置信度为70%。分类结果将结合本地物种信息进行人工比对, 当注释物种非当地物种时, 记为涵盖当地最近缘物种的最小分类单元, 最终序列物种注释到“目”水平。
物种注释完成后, 删除未鉴定成功序列以及那些不太可能成为欧亚水獭猎物的物种序列, 包括人类、欧亚水獭、细菌、真菌、藻类、水生微型动物(包括轮虫、缓步动物、溞类)以及小型环节动物(包括带丝蚓目、颤蚓目)的序列。此外当一个样品中某ASV序列数少于同批次PCR空白对照或少于该样品总序列数的1%时, 认为该序列可能来源于交叉污染, 舍弃这一序列, 该ASV不参与相应样品的食物结果与计算中。最后统计经筛选后的每份样品的物种类别以及对应序列条数用于食性数据分析。
1.5 食性数据分析
筛选后数据分析结果以相对出现频率(relative frequency of occurrence, %RFO)和相对序列丰度(relative read abundance, %RRA)两个指标(Deagle et al, 2019)表示。
%RFO是某一物种类别在所有样品中的出现次数占所有物种类别总出现次数的百分比, 计算公式如下:
其中, S为有效样品总数, T为物种类别总数, Ii,k为指示函数, 当物种类别i在样品k中存在其值记为1, 否则记为0。由公式可知, 全部物种类别的%RFO之和为100%。
%RRA是某一物种类别的序列数占该样品总物种类别序列的百分比, 反映了相对生物量, 计算公式如下:
其中, N为有效样品总数, T为物种类别总数, ni,k是物种类别i在样品k中的序列数。由公式可知, 全部物种类别的%RRA之和为100%。
为分析结果中不同物种类别的差异水平, 使用SPSS (26.0)软件中的独立样本Kruskal-Wallis H检验, 分别比较不同物种类别间的%RFO差异以及%RRA差异。
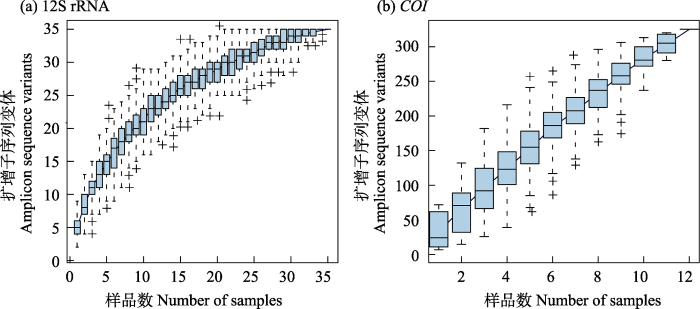
为评估样品数是否足够代表欧亚水獭食性结果, 使用R (4.1.2)中的vegan包(2.5-6)对经筛选后获得的物种ASV构建物种累积曲线。
2 结果
2.1 物种鉴定与高通量测序
本研究于大兴安岭北部4个研究区中共采集欧亚水獭肌肉样品1份及疑似水獭粪便样品50份。除5份DNA提取质量不合格及10份未扩增出目标条带外, 有35份粪便样品扩增出目标条带, 鉴定来源为欧亚水獭, 其中盘中20份, 呼中6份, 克一河5份, 甘河4份(附录1)。
对上述35个已鉴定样品进行食性分析, 其中12S rRNA区域全部扩增成功, COI区域仅成功扩增15份。测序结果经过滤、降噪、合并等处理后, 12S rRNA区域共获得80-120 bp双端序列片段5,855,222条, 平均每份样品167,292条序列(22,288-800,671), 其中1.22%来自欧亚水獭和人类, 另有3.83%低于1%阈值, 其余94.95%用于脊椎动物食性分析; COI区域共获得300-350 bp双端片段326,935条, 平均每份样品21,796条序列(1,294-40,396), 其中37.32%来自细菌、真菌、植物以及藻类, 19.85%未鉴定到目水平, 另有1.03%来自小型环节动物, 0.58%来自水生微型动物, 5.37%低于1%阈值, 其余35.84%用于后续无脊椎动物食性分析。
2.2 欧亚水獭食性分析
2.2.1 12S rRNA区域
处理结果显示, 12S rRNA区域有效样品数为35 (附录2), 序列注释筛选后共获得17种不同的物种类别, 其中11种鉴定到种水平, 6种鉴定到属水平(表2)。所有物种类别中, %RFO及%RRA大于10%的类别有杂色杜父鱼(Cottus poecilopus)以及黑龙江林蛙(Rana asmurensis), 其中杂色杜父鱼相对出现频率和相对序列丰度均最高, 分别为19.35%和27.32% (表2)。科水平上, 杜父鱼科、蛙科以及鲤科物种的相对出现频率和相对序列丰度均大于10%, 其中杜父鱼科均最高, 分别为32.26%和45.72% (图2)。从生物类群看, 相比蛙类, 17种物种类别中, 鱼类有15种, 物种最丰富(表2); 其次鱼类的%RFO和%RRA分别为81.94%和71.06%, 在脊椎动物物种类别中占主导地位(图2)。Kruskal-Wallis H检验结果表明, 科水平上, 不同物种类别在%RFO及%RRA统计值上存在显著差异(H = 107.08, df = 7)。类别的成对比较结果显示, 杜父鱼科与其他鱼类间具有显著差异。脊椎动物物种累积曲线显示, 随着样品数量的增加, 观测到的脊椎动物ASV数量趋于稳定, 说明本研究样品数量足以代表大兴安岭北部冬季欧亚水獭粪便中脊椎动物情况(图3a)。
表2 大兴安岭北部欧亚水獭粪便(N = 35) 12S rRNA区域食性分析结果
Table 2
| 物种类别 Species taxa | 出现样品数 Occurrence | 相对出现频率 %RFO | 相对序列丰度 %RRA | 生物类群 Biome |
|---|---|---|---|---|
| 杂色杜父鱼 Cottus poecilopus | 30 | 19.35 | 27.32 | 鱼类 Fish |
| 黑龙江林蛙 Rana amurensis | 24 | 15.48 | 21.73 | 蛙类 Frog |
| 克氏杜父鱼 Cottus czerskii | 15 | 9.68 | 16.47 | 鱼类 Fish |
| 东北林蛙 Rana dybowskii | 4 | 2.58 | 7.21 | 蛙类 Frog |
| 须鳅属一种 Barbatula sp. | 13 | 8.39 | 5.47 | 鱼类 Fish |
| 真鱥 Phoxinus phoxinus | 18 | 11.61 | 5.09 | 鱼类 Fish |
| 湖大吻鱥 Rhynchocypris percnurus | 10 | 6.45 | 4.65 | 鱼类 Fish |
| 黑龙江茴鱼 Thymallus arcticus | 8 | 5.16 | 2.96 | 鱼类 Fish |
| 拉氏大吻鱥 Rhynchocypris lagowskii | 8 | 5.16 | 2.81 | 鱼类 Fish |
| 黑龙江中杜父鱼 Mesocottus haitej | 2 | 1.29 | 1.54 | 鱼类 Fish |
| 茴鱼属一种 Thymallus sp. | 4 | 2.58 | 1.43 | 鱼类 Fish |
| 七鳃鳗属一种 Lampetra sp. | 4 | 2.58 | 1.08 | 鱼类 Fish |
| 葛氏鲈塘鳢 Perccottus glenii | 5 | 3.23 | 0.81 | 鱼类 Fish |
| 江鳕 Lota lota | 4 | 2.58 | 0.66 | 鱼类 Fish |
| 杜父鱼属一种 Cottus sp. | 3 | 1.94 | 0.39 | 鱼类 Fish |
| 大吻鱥属一种 Rhynchocypris sp. | 2 | 1.29 | 0.33 | 鱼类 Fish |
| 花鳅属一种 Cobitis sp. | 1 | 0.65 | 0.05 | 鱼类 Fish |
图2
图2
科水平上大兴安岭北部欧亚水獭粪便(N = 35)脊椎动物组成。(a)相对出现频率; (b)相对序列丰度。
Fig. 2
Vertebrate of Eurasian otter feces (N = 35) in northern Greater Khingan Mountains at family level. (a) Relative frequency of occurrence; (b) Relative read abundance.
图3
图3
12S rRNA和COI区域食性分析结果物种累积曲线
Fig. 3
Species accumulation curve of diet analysis in 12S rRNA and COI region
2.2.2 COI区域
处理结果显示, COI区域有效样品数为12 (附录3), 注释后共获得5种分类到目水平的物种类别。从生物类群看, 5种类别均为昆虫。所有类别中, %RFO及%RRA大于10%的有蜻蜓目、毛翅目、襀翅目和蜉蝣目(表3), 其中蜻蜓目(%RFO = 20.51%, %RRA = 33.86%)是粪便中无脊椎动物的主要组成部分。使用独立样本Kruskal-Wallis H检验比较不同目间的%RFO及%RRA统计值差异, 结果表明5个目水平的物种类别在%RFO及%RRA统计值上不存在显著差异。无脊椎动物物种累积曲线显示, 随着样品数量的增加, 观测到的无脊椎动物物种ASV数量仍呈上升趋势, 说明本研究仍需更多的样品数量以分析粪便中无脊椎动物的组成(图3b)。
表3 大兴安岭北部欧亚水獭粪便(N = 12) COI区域食性分析结果
Table 3
| 物种类别 Species taxa | 出现样品数 Occurrence | 相对出现频率 %RFO | 相对序列丰度 %RRA | 生物类群 Biome |
|---|---|---|---|---|
| 蜻蜓目 Odonata | 8 | 20.51 | 33.86 | 昆虫 Insect |
| 毛翅目 Trichoptera | 9 | 23.08 | 25.92 | 昆虫 Insect |
| 襀翅目 Plecoptera | 7 | 17.95 | 18.74 | 昆虫 Insect |
| 蜉蝣目 Ephemeroptera | 8 | 20.51 | 15.87 | 昆虫 Insect |
| 双翅目 Diptera | 7 | 17.95 | 5.60 | 昆虫 Insect |
3 讨论
3.1 物种鉴定
欧亚水獭一般独居, 多在夜间活动, 成年雄性活动范围可达15 km, 行动迅速, 行踪隐蔽, 难以被人直接观察(Erlinge, 1967; 韩雪松等, 2021)。同时从当年9月下旬到翌年4月上旬, 采样区域日平均气温均低于10℃, 极端最低气温可达-40℃, 人为长期监测难度大, 红外相机等长期监测设备受极端低温影响常无法正常工作, 因此难以通过直接观察其捕食行为获得食性数据。得益于分子生物学的发展, 目前基于粪便DNA获取食物组成已成为食性研究的主流方法。然而该方法需要物种来源明确的粪便样品作为研究材料, 以避免得出错误的食性结果(Morin et al, 2016)。但基于粪便形态特征采集样品往往有误判来源的风险, 粪便质量也无法得到保证, 因此在食性分析前对样品的质量及物种来源进行检测就显得尤为重要(陆琪等, 2019; Harper et al, 2020)。
本研究于大兴安岭北部共采集疑似水獭粪便样品50份, 在采集时基于欧亚水獭习性, 根据雪地痕迹以及粪便形态特征预判为欧亚水獭粪便, 除5份DNA提取质量不合格及10份未扩增出目标条带外, 剩余35份均成功扩增出目标条带被鉴定源自欧亚水獭, 有效样品概率为70%。本次实验中粪便物种鉴定阶段所使用的引物为针对欧亚水獭设计的特异引物LLCBL/R, 该引物已被成功应用于淡水生态系统中欧亚水獭的检测(Thomsen et al, 2012)。同时在后续食性分析阶段对脊椎动物12S rRNA区域进行测序后, 35份样品中均存在欧亚水獭的序列且无其他捕食者序列, 说明该物种鉴定方法较为可靠。
本次实验中较低的鉴定成功率可能是因为采集的样品质量不佳或保存方法不当导致了DNA的降解。目前较新的基于粪便DNA的动物食性研究一般采用两步法保存粪便, 即先用无水乙醇或95%酒精浸泡24 h后, 再加入硅胶进行-20℃保存(Shao et al, 2021; Wang et al, 2022)。有研究表明, 采用两步法要比直接加入硅胶, 能从粪便中提取到更高浓度的DNA, 在PCR实验中也有更高的成功率(Nsubuga et al, 2004)。因此, 本研究中直接加入硅胶进行低温保存可能会对DNA提取和扩增的效果造成影响, 使鉴定成功率降低。同时需要强调的是, 尽管后续实验结果表明使用硅胶冷冻法保存的样品仍可以用于食性分析, 但由于研究区域和季节的特殊性, 本研究采样时日平均气温均在-10℃以下, 而低温环境有利于粪便DNA的保存。因此本研究中使用的粪便保存方法可能不适用于其他地区或其他季节。另一个导致成功率低的因素可能是采集的粪便来源于非欧亚水獭物种, 因此无法扩增到目标条带。上述问题可通过尽量采集新鲜粪便及优化保存方法以减少降解所带来的影响。
3.2 食性分析
传统的基于形态学的欧亚水獭食性研究受限于食物残渣的辨识度, 往往只能将猎物鉴定到较高的分类水平, 同时由于同源物种间形态相近容易误判, 因此难以在物种水平上获得准确的食性结果(Britton et al, 2006; Smiroldo et al, 2019; Marcolin et al, 2020)。利用DNA宏条形码技术, 本研究成功从欧亚水獭粪便中鉴定出分类到种的脊椎动物共计11种及分类到目的无脊椎动物共计5种, 其中脊椎动物分类精度与同样使用DNA宏条形码的食性研究相当(Harper et al, 2020; Wang et al, 2022)。更精细的食性结果, 说明了DNA宏条形码技术可用于欧亚水獭的食性研究且具有更大的应用潜力。
本研究使用了两组引物分别探究欧亚水獭对脊椎动物及无脊椎动物的摄食情况, 尽管基于高通量测序的食性结果可以用于半定量分析, 但由于不同引物间的扩增成功率与效率存在差异, 序列丰度与生物量之间的关系往往会发生变化, 且相关性较弱, 因此本研究未对两组引物扩增获得的食性结果进行统一分析(Pompanon et al, 2012; Kumari et al, 2019)。已有研究表明, 欧亚水獭作为机会主义捕食者, 其食谱组成受到当地食物资源多样性影响, 但主要以鱼类为主, 其次是两栖动物、鸟类、爬行动物和无脊椎动物(Clavero et al, 2003; Krawczyk et al, 2016; Lanszki et al, 2016)。同时尽管欧亚水獭食性在不同季节、不同栖息地类型、不同地理区域中存在差异, 但鱼类仍是水獭食物的主要来源, 而两栖动物常在特定的季节(如冬春季)或特定的水体(如小溪沼泽)中作为鱼类的重要食物补充(Taastrøm & Jacobsen, 1999; Smiroldo et al, 2009; Krawczyk et al, 2016; Lanszki et al, 2016)。此外有研究表明, 昆虫在水獭粪便中出现频率较高, 但只占其食物生物量的很小一部分, 食物重要性远小于鱼类、蛙类等脊椎动物(Krawczyk et al, 2016)。因此尽管本研究对两组引物扩增结果分别进行分析, 但仍将鱼类、蛙类等脊椎动物作为主要食物来源进行讨论。
对脊椎动物12S rRNA区域序列的分析结果表明, 大兴安岭北部欧亚水獭冬季主要捕食鱼类(%RFO = 81.94%, %RAA = 71.06%), 其次是蛙类(%RFO = 18.06%, %RAA = 28.94%), 而不捕食鸟类、爬行动物及小型哺乳动物。这一结果与欧洲的欧亚水獭食性研究结果相似, 其食物组成都是以鱼类为主, 其次为两栖动物, 不同的是在欧洲的相关研究中水獭还会捕食鸟类、小型哺乳动物等脊椎动物(Clavero et al, 2003; Krawczyk et al, 2016; Lanszki et al, 2016)。例如, Harper等(2020)在英国北部和东部基于DNA宏条形码技术开展欧亚水獭食性研究, 发现水獭的食物主要是鱼类(81.1%), 其次是两栖动物(12.7%)、鸟类(5.9%)和哺乳动物(0.5%)。国内的欧亚水獭食性研究表明, 水獭主要捕食鱼类, 其次是两栖动物、鸟类和哺乳动物, 而不食用爬行动物(Wang et al, 2022)。本研究中, 蛙类是除鱼类外唯一对饮食做出重要贡献的脊椎动物猎物。这可能是冬季时蛙类冬眠, 相较鸟类、爬行动物、哺乳动物等物种更容易被捕捉(Weber, 1990)。此外, 由于冬季研究区河流已经封冻, 极端天气下, 营水边的鸟类数量减少也可能导致结果未能扩增出鸟类序列。还需注意的是, 有研究表明, 本研究所使用的通用脊椎动物引物12S V5并不是对所有脊椎动物都有效(Kelly et al, 2014), 因此还需要考虑引物是否充分覆盖研究区潜在食源物种做进一步研究。
本研究中欧亚水獭较多捕食杂色杜父鱼、克氏杜父鱼(Cottus czerskii)等杜父鱼科鱼类(%RRA = 45.72%), 而较少捕食其他鱼类。这可能是由于欧亚水獭偏好捕食移动缓慢和体型较小的猎物。杂色杜父鱼、克氏杜父鱼等杜父鱼科鱼类在形态上表现为小型鱼类, 主要栖息于淡水水系中较为复杂的山涧及其支流, 常被发现生存于水温较低的砂砾与石缝中, 活动能力较弱, 是典型的山涧冷水性底层鱼类①(①周远帆 (2021) 东北地区杂色杜父鱼线粒体全基因组序列测定及遗传多样性分析. 硕士学位论文, 上海海洋大学, 上海.)。而其他鱼类如湖大吻鱥(Rhynchocypris percnurus)、真鱥(Phoxinus phoxinu)等也属于冷水性小型鱼类(许旺, 2013), 但其活动能力较强, 捕捉它们需要更多的能量与时间。与本研究中水獭鱼类捕食倾向相似, 以往有研究表明, 欧亚水獭更倾向于捕食移动缓慢和较小的底栖鱼类(Hong et al, 2019; Harper et al, 2020)。但也有研究指出, 欧亚水獭更喜欢捕食能快速活动的鲤科鱼类(Kumari et al, 2019; Wang et al, 2022)。研究结果的差异可能与栖息地猎物的捕捉难度密切相关, 而捕捉难度主要受栖息地猎物组成、水体类型、季节等因素的影响(Wang et al, 2022)。因此本研究得出的欧亚水獭鱼类偏好性的结论仍需结合当地的物种情况, 在不同栖息地类型、不同季节做进一步研究。
除了使用针对脊椎动物的12S V5引物外, 为了探讨欧亚水獭捕食无脊椎动物的可能, 本研究还使用了针对无脊椎动物的BF2/BR1引物对粪便DNA进行扩增。结果发现除了水生微型动物、小型环节动物、昆虫等无脊椎动物外, 还扩增出了细菌、真菌、藻类以及植物等非无脊椎动物序列。非无脊椎动物序列的产生可能是由于使用引物的简并性过高引起的非特异性扩增。此外植物、藻类、水生微型动物(轮虫、溞类等)以及小型环节动物(包括带丝蚓目、颤蚓目)序列的产生可能是作为鱼类食物被一同扩增使结果产生了偏差(Alberdi et al, 2019)。因此根据以往的食性研究成果, 本研究删除了那些不太可能成为欧亚水獭猎物的物种序列进行分析。
对无脊椎动物COI区域序列的分析结果表明, 欧亚水獭粪便中存在蜻蜓目、毛翅目、襀翅目、蜉蝣目以及双翅目等昆虫, 且无其他大型无脊椎动物的存在, 这表明研究区水獭有可能捕食昆虫作为食物补充。但昆虫作为鱼类的常见食物之一, 极有可能随着水獭捕食鱼类而出现在水獭粪便中。有研究表明, 毛翅目、襀翅目、蜉蝣目等昆虫是杂色杜父鱼的主要食物来源(霍堂斌等, 2015)。且基于显著性差异分析发现, 5个昆虫类别中, 小型昆虫(难以被水獭主动捕食)如双翅目和毛翅目昆虫与大型昆虫(易被水獭发现捕食)如蜻蜓目在%RFO及%RRA统计值上不存在显著差异。这表明样品中的昆虫很有可能来源于鱼类或者蛙类等被捕食者体内。因此尽管已有研究发现, 欧亚水獭在圈养情况下会积极捕食大型昆虫(Carss & Parkinson, 1996), 且相关食性研究表明, 昆虫一直是水獭无脊椎动物食物的重要组成部分(Krawczyk et al, 2016; Lanszki et al, 2016)。但出于保守性原则, 本研究不将蜻蜓目、蜉蝣目等昆虫列入大兴安岭北部欧亚水獭冬季的食性中。此外本研究并未从水獭粪便中发现虾类、蟹类、蚌类等大型无脊椎动物的存在, 这可能与栖息地相关物种数量稀少有关(经访谈, 研究区管护人员表示很少在研究区河段中发现虾类、蟹类、蚌类等大型无脊椎动物)。
附录 Supplementary Material
附录1 经物种鉴定筛选出的有效欧亚水獭粪便样品
Appendix 1 The valid fecal samples of Eurasian otter screened out through DNA identification in the northern Greater Khingan Mountains
附录2 科水平上大兴安岭北部欧亚水獭粪便(N = 35) 12S rRNA区域物种注释结果
Appendix 2 Results of diet analysis at family level in 12S rRNA region (N = 35) of Eurasian otter fecal samples in the northern Greater Khingan Mountains
附录3 目水平上大兴安岭北部欧亚水獭粪便(N = 12) COI区域物种注释结果
Appendix 3 Results of diet analysis at order level in COI region (N = 12) of Eurasian otter fecal samples in the northern Greater Khingan Mountains
参考文献
Promises and pitfalls of using high-throughput sequencing for diet analysis
DOI:10.1111/1755-0998.12960
PMID:30358108
[本文引用: 1]

The application of high-throughput sequencing-based approaches to DNA extracted from environmental samples such as gut contents and faeces has become a popular tool for studying dietary habits of animals. Due to the high resolution and prey detection capacity they provide, both metabarcoding and shotgun sequencing are increasingly used to address ecological questions grounded in dietary relationships. Despite their great promise in this context, recent research has unveiled how a wealth of biological (related to the study system) and technical (related to the methodology) factors can distort the signal of taxonomic composition and diversity. Here, we review these studies in the light of high-throughput sequencing-based assessment of trophic interactions. We address how the study design can account for distortion factors, and how acknowledging limitations and biases inherent to sequencing-based diet analyses are essential for obtaining reliable results, thus drawing appropriate conclusions. Furthermore, we suggest strategies to minimize the effect of distortion factors, measures to increase reproducibility, replicability and comparability of studies, and options to scale up DNA sequencing-based diet analyses. In doing so, we aim to aid end-users in designing reliable diet studies by informing them about the complexity and limitations of DNA sequencing-based diet analyses, and encourage researchers to create and improve tools that will eventually drive this field to its maturity.© 2018 John Wiley & Sons Ltd.
q2-sample-classifier: Machine-learning tools for microbiome classification and regression
Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2
Revealing the prey items of the otter Lutra lutra in South West England using stomach contents analysis
DADA2: High-resolution sample inference from Illumina amplicon data
DOI:10.1038/nmeth.3869
PMID:27214047
[本文引用: 1]

We present the open-source software package DADA2 for modeling and correcting Illumina-sequenced amplicon errors (https://github.com/benjjneb/dada2). DADA2 infers sample sequences exactly and resolves differences of as little as 1 nucleotide. In several mock communities, DADA2 identified more real variants and output fewer spurious sequences than other methods. We applied DADA2 to vaginal samples from a cohort of pregnant women, revealing a diversity of previously undetected Lactobacillus crispatus variants.
Errors associated with otter Lutra lutra faecal analysis. I. Assessing general diet from spraints
DOI:10.1111/jzo.1996.238.issue-2 URL [本文引用: 2]
Trophic diversity of the otter (Lutra lutra L.) in temperate and Mediterranean freshwater habitats
DOI:10.1046/j.1365-2699.2003.00865.x URL [本文引用: 2]
DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: Application to omnivorous diet
DOI:10.1111/1755-0998.12188
PMID:24128180
[本文引用: 1]

Ecological understanding of the role of consumer-resource interactions in natural food webs is limited by the difficulty of accurately and efficiently determining the complex variety of food types animals have eaten in the field. We developed a method based on DNA metabarcoding multiplexing and next-generation sequencing to uncover different taxonomic groups of organisms from complex diet samples. We validated this approach on 91 faeces of a large omnivorous mammal, the brown bear, using DNA metabarcoding markers targeting the plant, vertebrate and invertebrate components of the diet. We included internal controls in the experiments and performed PCR replication for accuracy validation in postsequencing data analysis. Using our multiplexing strategy, we significantly simplified the experimental procedure and accurately and concurrently identified different prey DNA corresponding to the targeted taxonomic groups, with ≥ 60% of taxa of all diet components identified to genus/species level. The systematic application of internal controls and replication was a useful and simple way to evaluate the performance of our experimental procedure, standardize the selection of sequence filtering parameters for each marker data and validate the accuracy of the results. Our general approach can be adapted to the analysis of dietary samples of various predator species in different ecosystems, for a number of conservation and ecological applications entailing large-scale population level diet assessment through cost-effective screening of multiple DNA metabarcodes, and the detection of fine dietary variation among samples or individuals and of rare food items.© 2013 John Wiley & Sons Ltd.
Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data?
DOI:10.1111/mec.14734
PMID:29858539
[本文引用: 1]

Advances in DNA sequencing technology have revolutionized the field of molecular analysis of trophic interactions, and it is now possible to recover counts of food DNA sequences from a wide range of dietary samples. But what do these counts mean? To obtain an accurate estimate of a consumer's diet should we work strictly with data sets summarizing frequency of occurrence of different food taxa, or is it possible to use relative number of sequences? Both approaches are applied to obtain semi-quantitative diet summaries, but occurrence data are often promoted as a more conservative and reliable option due to taxa-specific biases in recovery of sequences. We explore representative dietary metabarcoding data sets and point out that diet summaries based on occurrence data often overestimate the importance of food consumed in small quantities (potentially including low-level contaminants) and are sensitive to the count threshold used to define an occurrence. Our simulations indicate that using relative read abundance (RRA) information often provides a more accurate view of population-level diet even with moderate recovery biases incorporated; however, RRA summaries are sensitive to recovery biases impacting common diet taxa. Both approaches are more accurate when the mean number of food taxa in samples is small. The ideas presented here highlight the need to consider all sources of bias and to justify the methods used to interpret count data in dietary metabarcoding studies. We encourage researchers to continue addressing methodological challenges and acknowledge unanswered questions to help spur future investigations in this rapidly developing area of research.© 2018 John Wiley & Sons Ltd.
Impacts of Changes in Climate, Forest Cover and Permafrost on Streamflow in the Da Hinggan Mountains watersheds
大兴安岭气候、森林覆盖率和冻土变化对河川径流的影响
The functional role of biodiversity in ecosystems: Incorporating trophic complexity
Understanding how biodiversity affects functioning of ecosystems requires integrating diversity within trophic levels (horizontal diversity) and across trophic levels (vertical diversity, including food chain length and omnivory). We review theoretical and experimental progress toward this goal. Generally, experiments show that biomass and resource use increase similarly with horizontal diversity of either producers or consumers. Among prey, higher diversity often increases resistance to predation, due to increased probability of including inedible species and reduced efficiency of specialist predators confronted with diverse prey. Among predators, changing diversity can cascade to affect plant biomass, but the strength and sign of this effect depend on the degree of omnivory and prey behaviour. Horizontal and vertical diversity also interact: adding a trophic level can qualitatively change diversity effects at adjacent levels. Multitrophic interactions produce a richer variety of diversity-functioning relationships than the monotonic changes predicted for single trophic levels. This complexity depends on the degree of consumer dietary generalism, trade-offs between competitive ability and resistance to predation, intraguild predation and openness to migration. Although complementarity and selection effects occur in both animals and plants, few studies have conclusively documented the mechanisms mediating diversity effects. Understanding how biodiversity affects functioning of complex ecosystems will benefit from integrating theory and experiments with simulations and network-based approaches.
Validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment
Home range of the otter Lutra lutra L. in southern Sweden
DOI:10.2307/3565098 URL [本文引用: 1]
Illegal Otter Trade: An Analysis of Seizures in Selected Asian Countries (1980-2015)
Using surveillance cameras to analyze the activity pattern of the Eurasian otters (Lutra lutra) and the efficiency of camera trap monitoring
DOI:10.17520/biods.2020388 URL [本文引用: 2]
基于视频监控系统的欧亚水獭活动节律初报及红外相机监测效果评估
Using DNA metabarcoding to investigate diet and niche partitioning in the native European otter (Lutra lutra) and invasive American mink (Neovison vison)
DOI:10.3897/mbmg.4.56087
URL
[本文引用: 5]

In the UK, the native European otter (Lutra lutra) and invasive American mink (Neovison vison) have experienced concurrent declines and expansions. Currently, the otter is recovering from persecution and waterway pollution, whereas the mink is in decline due to population control and probable interspecific interaction with the otter. We explored the potential of DNA metabarcoding for investigating diet and niche partitioning between these mustelids. Otter spraints (n = 171) and mink scats (n = 19) collected from three sites (Malham Tarn, River Hull and River Glaven) in northern and eastern England were screened for vertebrates using high-throughput sequencing. Otter diet mainly comprised aquatic fishes (81.0%) and amphibians (12.7%), whereas mink diet predominantly consisted of terrestrial birds (55.9%) and mammals (39.6%). The mink used a lower proportion (20%) of available prey (n = 40 taxa) than the otter and low niche overlap (0.267) was observed between these mustelids. Prey taxon richness of mink scats was lower than otter spraints and beta diversity of prey communities was driven by taxon turnover (i.e. the otter and mink consumed different prey taxa). Considering otter diet only, prey taxon richness was higher in spraints from the River Hull catchment and beta diversity of prey communities was driven by taxon turnover (i.e. the otter consumed different prey taxa at each site). Studies using morphological faecal analysis may misidentify the predator as well as prey items. Faecal DNA metabarcoding can resolve these issues and provide more accurate and detailed dietary information. When scaled up across multiple habitat types, DNA metabarcoding should greatly improve future understanding of resource use and niche overlap between the otter and mink.
A molecular approach to identifying the relationship between resource use and availability in Eurasian otters (Lutra lutra)
DOI:10.1139/cjz-2018-0289
URL
[本文引用: 1]

In South Korea, the Eurasian otter (Lutra lutra (Linnaeus, 1758)), a semi-aquatic carnivore, is found mainly in lower order streams that tend to have a low abundance of preferred prey fish species. To investigate the relationship between resource use and availability, we used DNA barcoding to identify otter diet items in 24 otter spraints (faeces) from 16 sites along the Nakdong River basin from 4 to 6 June 2014. At these sites fish availability was assessed using scoop nets and casting nets. Fish formed the bulk of otter diet, which included also frogs, mammals, and reptiles. By DNA barcoding (success rate: 72.38%), we identified 79 prey items from 105 bone remains. The diet comprised mostly fish, but frogs, mammals, and reptiles were also identified. The fish fauna and otter diet composition differed significantly. Across the study sites, members of the Cyprinidae dominated in netted samples, but occurred less frequently in otter diet. Because most Cyprinidae are fast swimmers, otters also fed on benthic fishes and frogs, suggesting limited foraging flexibility in otters and specialization on more slowly moving prey.
Biological characteristics of alpine sculpin Cottus poecilopus Heckel
杂色杜父鱼生物学初步研究
DNA metabarcoding illuminates dietary niche partitioning by African large herbivores
Using environmental DNA to census marine fishes in a large mesocosm
DOI:10.1371/journal.pone.0086175 URL [本文引用: 1]
A comparison and critique of different scat-analysis methods for determining carnivore diet
DOI:10.1111/mam.2011.41.issue-4 URL [本文引用: 2]
Diet composition of the Eurasian otter Lutra lutra in different freshwater habitats of temperate Europe: A review and meta-analysis
DOI:10.1111/mam.12054 URL [本文引用: 5]
Otters:Ecology
DNA metabarcoding-based diet survey for the Eurasian otter (Lutra lutra): Development of a Eurasian otter-specific blocking oligonucleotide for 12S rRNA gene sequencing for vertebrates
Diet of otters (Lutra lutra) in various habitat types in the Pannonian biogeographical region compared to other regions of Europe
DOI:10.7717/peerj.2266
URL
[本文引用: 4]

Knowledge of the effect of habitat type and region on diet and feeding behaviours of a species facilitates a better understanding of factors impacting populations, which contributes to effective conservation management. Using spraint analysis and relative frequency of occurrence data from the literature, we described the dietary patterns of Eurasian otters (Lutra lutra) in 23 study sites within the Pannonian biogeographical region in Hungary. Our results indicated that diet composition varied by habitat type and is therefore context dependant. The differences among habitat types were however lower than expected. We noticed a decline in the fish consumption with a concomitant increase in trophic niche breadth and amphibian consumption in rivers, ponds (fish farms), backwaters, marshes and small watercourses. The main differences in diet were not attributed to the consumption of primary and secondary food types (fish and amphibians), but rather to differences in other, less important food types (mammals, birds). Using hierarchical cluster analysis, rivers and ponds could clearly be separated from other habitat types. We found the main fish diet of otters in most of these areas consisted of small (<100 g), eurytopic, littoral and non-native, mostly invasive species. Dietary studies from 91 sites in six European biogeographical regions showed that fish are consumed most frequently in the Atlantic and Boreal, less in the Continental and Pannonian, and least in the Alpine and Mediterranean regions. Comparative analysis indicated that the Mediterranean region (with frequent crayfish consumption) and Alpine region (frequent amphibian consumption) cluster separate from the other regions.
Past and present: The status and distribution of otters (Carnivora: Lutrinae) in China
DOI:10.1017/S0030605317000400
URL
[本文引用: 1]

Three species of otters are known from China; the Eurasian otter Lutra lutra is widespread throughout the country and the smooth-coated Lutrogale perspicillata and Asian small-clawed otters Aonyx cinereus occur in tropical and subtropical regions. We summarize the past status and distribution of otters in China, and provide an update based on a literature review, interviews and field surveys. Otter populations have undergone a dramatic countrywide decline, and are extirpated over much of their former ranges. Relict populations persist, however, in well-protected nature reserves, in sparsely populated headwaters of the Qinghai–Tibetan Plateau, at remote sites along international borders, and in densely populated deltas and floodplains. Recent records were mostly of the Eurasian otter, and we could find no confirmed recent record of the smooth-coated otter. The otters that survive in certain well-protected sites could act as source populations for recolonization if adequate conservation interventions are implemented. Urgent, focused action is needed to protect the remaining populations, and to study the taxonomy and ecology of China's otters.
Community structure and bioindicator of benthic fauna from Huma to Heihe in the upper reaches of Heilongjiang Province
黑龙江上游呼玛至黑河江段底栖动物群落结构及其生物指示作用
Potential distribution of otter in Northeast China
中国东北地区水獭种群潜在分布区的预测
Metabarcoding diet analysis of snow leopards (Panthera uncia) in Wolong National Nature Reserve, Sichuan Province
DOI:10.17520/biods.2019101
[本文引用: 2]

As the apex predator of plateau ecosystems in Central Asia and the Qinghai-Tibet Plateau, the snow leopard (Panthera uncia) plays an essential role in maintaining food-web structure and ecosystem stability. Learning the diet composition and dynamics of the snow leopard is important for understanding its role in ecosystem functioning and interspecific interactions. Previous diet analyses of the snow leopard have been based mainly on morphological identification of food debris in the feces, though the accuracy of this practice has been broadly debated. The Qionglai Mountains are located at the southeast edge of the snow leopard range, harboring a small and relatively isolated population of snow leopards that are barely studied. Using non-invasive sampling, we collected 38 putative snow leopard fecal samples in the Wolong National Nature Reserve in the Qionglai Mountains. To identify the fecal origin, we extracted the fecal DNA and amplified the mitochondrial DNA 16S rRNA gene fragment. Twenty-two fecal samples were identified as originating from snow leopards. Subsequently, vertebrate universal primers and a snow leopard-specific blocking oligo were used to amplify the food components in the fecal DNA, and then high-throughput sequencing was performed to analyze the diet composition of snow leopards. The blue sheep (Pseudois nayaur) was detected in 67% of the samples and was found to be the main staple food of snow leopards’ diet. The domestic yak (Bos grunniens) appeared in 33% of the fecal samples, also accounting for a high proportion of the snow leopard diet. In addition, pikas (Ochotona spp.) and birds were found in a small number of fecal samples. Therefore, wild prey was found to be the main food source for snow leopards in Wolong. However, livestock (yak) also accounted for a relatively large proportion of their diet.
基于分子宏条形码分析四川卧龙国家级自然保护区雪豹的食性
DOI:10.17520/biods.2019101
[本文引用: 2]

作为中亚和青藏高原山地生态系统中的顶级捕食者, 雪豹(Panthera uncia)对于维持食物网结构和生态系统稳定性有重要作用。了解雪豹的食性组成和变化对于理解其生态系统功能和物种间相互作用有重要意义。以往的雪豹食性分析多基于对其粪便中食物残渣的形态学鉴定, 但准确度受人员经验和主观因素影响较大。邛崃山脉位于雪豹分布区东南缘, 该区域的雪豹种群规模小且相对孤立, 研究匮乏。本研究基于非损伤性取样, 在邛崃山脉的卧龙国家级保护区采集疑似雪豹粪便样品38份, 首先提取粪便DNA, 并扩增线粒体DNA 16S rRNA基因片段进行分子物种鉴定, 确定其中22份为雪豹粪便样品。随后, 利用脊椎动物通用引物和雪豹特异性阻抑引物扩增粪便DNA中的食物成分, 并进行高通量测序, 分析雪豹食性构成。食性分析结果显示岩羊(Pseudois nayaur)是卧龙地区雪豹最主要的食物, 在67%的样品中均有检出。家牦牛(Bos grunniens)在33%的样品中出现, 也在雪豹食性中占较高比例。此外, 鼠兔(Ochotona spp.)和鸟类也在少量样品中发现。可见, 野生猎物是卧龙地区雪豹的主要食物资源; 与大世界大多数其他地区的雪豹食性相同, 野生大型有蹄类是雪豹最重要的食物。然而家畜(牦牛)在卧龙雪豹食谱中有相当高的占比, 显示该区域内可能存在较为严重的由雪豹捕食散养家畜引起的人兽冲突问题。
Otter diet and prey selection in a recently recolonized area assessed using microscope analysis and DNA barcoding
Feeding ecological knowledge: The underutilised power of faecal DNA approaches for carnivore diet analysis
DOI:10.1111/mam.12144
[本文引用: 3]

Accurate analyses of the diets of predators are key to understand trophic interactions and defining conservation strategies. Diets are commonly assessed through analysis of non-invasively collected scats, and the use of faecal DNA (fDNA) analysis can reduce the species misidentifications that could lead to biased ecological inference. We review the scientific literature since publication of the first paper on amplifying fDNA, in order to assess trends in the use of genetic non-invasive sampling (gNIS) for predator species identification in scat-based diet studies of North American and European terrestrial mammalian carnivores (Carnivora). We quantify error rates in morphology-based predator species identification. We then provide an overview of how applying gNIS would improve research on trophic interactions and other areas of carnivore ecology. We found that carnivore species identity was verified by using gNIS in only 8% of 400 studies of carnivore diets based on scats. The median percentage of false positives (i.e. samples wrongly identified as belonging to the target species) in morphology-based studies was 18%, and was consistent regardless of species' body size. We did not find an increasing trend in the use of gNIS over time, despite the existing technical capability to identify almost all carnivore species. New directions for fDNA studies include employing high-throughput sequencing (HTS) and DNA metabarcoding to identify the predator species, the individual predator, the entire assemblage of consumed items, and the microbiome of the predator and pathogens. We conclude that HTS protocols and metagenomic approaches hold great promise for elevating gNIS as a fundamental cornerstone for future research in ecology and conservation biology of mammals.
Bias in carnivore diet analysis resulting from misclassification of predator scats based on field identification
DOI:10.1002/wsb.v40.4 URL [本文引用: 1]
Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method
DOI:10.1111/j.1365-294X.2004.02207.x
PMID:15189228
[本文引用: 1]

Genetic analysis using noninvasively collected samples such as faeces continues to pose a formidable challenge because of unpredictable variation in the extent to which usable DNA is obtained. We investigated the influence of multiple variables on the quantity of DNA extracted from faecal samples from wild mountain gorillas and chimpanzees. There was a small negative correlation between temperature at time of collection and the amount of DNA obtained. Storage of samples either in RNAlater solution or dried using silica gel beads produced similar results, but significantly higher amounts of DNA were obtained using a novel protocol that combines a short period of storage in ethanol with subsequent desiccation using silica.
Who is eating what: Diet assessment using next generation sequencing
DOI:10.1111/j.1365-294X.2011.05403.x
PMID:22171763
[本文引用: 2]

The analysis of food webs and their dynamics facilitates understanding of the mechanistic processes behind community ecology and ecosystem functions. Having accurate techniques for determining dietary ranges and components is critical for this endeavour. While visual analyses and early molecular approaches are highly labour intensive and often lack resolution, recent DNA-based approaches potentially provide more accurate methods for dietary studies. A suite of approaches have been used based on the identification of consumed species by characterization of DNA present in gut or faecal samples. In one approach, a standardized DNA region (DNA barcode) is PCR amplified, amplicons are sequenced and then compared to a reference database for identification. Initially, this involved sequencing clones from PCR products, and studies were limited in scale because of the costs and effort required. The recent development of next generation sequencing (NGS) has made this approach much more powerful, by allowing the direct characterization of dozens of samples with several thousand sequences per PCR product, and has the potential to reveal many consumed species simultaneously (DNA metabarcoding). Continual improvement of NGS technologies, on-going decreases in costs and current massive expansion of reference databases make this approach promising. Here we review the power and pitfalls of NGS diet methods. We present the critical factors to take into account when choosing or designing a suitable barcode. Then, we consider both technical and analytical aspects of NGS diet studies. Finally, we discuss the validation of data accuracy including the viability of producing quantitative data.© 2011 Blackwell Publishing Ltd.
ecoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis
DOI:10.1093/nar/gkr732 URL [本文引用: 2]
VSEARCH: A versatile open source tool for metagenomics
DOI:10.7717/peerj.2584
URL
[本文引用: 1]

VSEARCH is an open source and free of charge multithreaded 64-bit tool for processing and preparing metagenomics, genomics and population genomics nucleotide sequence data. It is designed as an alternative to the widely used USEARCH tool (Edgar, 2010) for which the source code is not publicly available, algorithm details are only rudimentarily described, and only a memory-confined 32-bit version is freely available for academic use.
Is the otter a bioindicator?
Variation in diet composition and its relation to gut microbiota in a passerine bird
DOI:10.1038/s41598-022-07672-9
PMID:35260644
[本文引用: 1]

Quality and quantity of food items consumed has a crucial effect on phenotypes. In addition to direct effects mediated by nutrient resources, an individual's diet can also affect the phenotype indirectly by altering its gut microbiota, a potent modulator of physiological, immunity and cognitive functions. However, most of our knowledge of diet-microbiota interactions is based on mammalian species, whereas little is still known about these effects in other vertebrates. We developed a metabarcoding procedure based on cytochrome c oxidase I high-throughput amplicon sequencing and applied it to describe diet composition in breeding colonies of an insectivorous bird, the barn swallow (Hirundo rustica). To identify putative diet-microbiota associations, we integrated the resulting diet profiles with an existing dataset for faecal microbiota in the same individual. Consistent with previous studies based on macroscopic analysis of diet composition, we found that Diptera, Hemiptera, Coleoptera and Hymenoptera were the dominant dietary components in our population. We revealed pronounced variation in diet consumed during the breeding season, along with significant differences between nearby breeding colonies. In addition, we found no difference in diet composition between adults and juveniles. Finally, our data revealed a correlation between diet and faecal microbiota composition, even after statistical control for environmental factors affecting both diet and microbiota variation. Our study suggests that variation in diet induce slight but significant microbiota changes in a non-mammalian host relying on a narrow spectrum of items consumed.© 2022. The Author(s).
The role of forage availability on diet choice and body condition in American beavers (Castor canadensis)
DOI:10.1016/j.mambio.2012.12.001 URL [本文引用: 1]
Generalist carnivores can be effective biodiversity samplers of terrestrial vertebrates
DOI:10.1002/fee.v19.10 URL [本文引用: 1]
Fast surveys and molecular diet analysis of carnivores based on fecal DNA and metabarcoding
DOI:10.17520/biods.2018214
[本文引用: 2]

Large carnivores play an important role in the regulation of food-web structure and ecosystem functioning. However, large carnivores face serious threats that have caused declines in their populations and geographic ranges due to habitat loss and degradation, hunting, human disturbance and pathogen transmission. Conservation of large carnivore species richness and population size has become a pressing issue and an important research focus of conservation biology. The western Sichuan Plateau, located at the intersection of the mountains of southwest China and the eastern margin of the Tibetan Plateau, is a global biodiversity hotspot and has high carnivore species richness. However, increasing human activities may exacerbate the destruction of local flora and fauna, thereby threatening the survival of wild carnivores. Information on species composition and dietary habits can improve our understanding of the structure and function of the ecosystem and food-web relationships in the study area. In addition, species composition and dietary habits are of great significance for understanding multi-species coexistence mechanisms and preserving biodiversity. This study collected carnivore fecal samples from Xinlong and Shiqu counties in the Ganzi Tibetan Autonomous Prefecture, Sichuan Province. DNA was then extracted from the samples and the species was identified based on DNA sequences and DNA barcoding techniques. Seven carnivores were identified, including five large carnivores (Canis lupus, Ursus arctos, Panthera pardus, P. uncia and Canis lupus familiaris) and two medium and small-sized carnivores (Prionailurus bengalensis and Vulpes vulpes). Using fecal DNA, high-throughput sequencing and metabarcoding, we conducted diet analysis for the seven carnivores and found 28 different food molecular operational taxonomic units (MOTUs), including 19 mammals, eight birds and one fish species. The predominant prey categories of wolves, dogs and brown bears were ungulates. The domestic yak (Bos grunniens) was the most frequently identified prey species. Small mammals such as rodents and lagomorphs accounted for a significant proportion in the diets of leopard cats and red foxes, The most frequent prey of this category of carnivore were the Chinese scrub vole (Neodon irene) and plateau pika (Ochotona curzoniae). In addition, leopards and snow leopards mainly fed on the Chinese goral (Naemorhedus griseus) and blue sheep (Pseudois nayaur), respectively. Our study highlights the utility of fecal DNA and metabarcoding technique in fast carnivore surveys and high-throughput diet analysis, and provides a technical reference and guidance for future biodiversity surveys and food-web studies.
基于粪便DNA及宏条形码技术的食肉动物快速调查及食性分析
DOI:10.17520/biods.2018214
[本文引用: 2]

在陆地生态系统中, 大型食肉动物对于稳定食物网结构和生态系统功能有重要作用。在世界范围内, 由于栖息地丧失和破碎化、猎杀、人类活动干扰以及病原体的传播, 大型食肉动物生存正面临严重威胁, 多种食肉动物地理分布范围及种群数量大幅度缩减。如何有效保护大型食肉动物物种多样性及种群已经成为世界关注的焦点问题和保护生物学的重要研究方向。川西高原地处我国西南山地与青藏高原东缘交界地带, 属于世界生物多样性热点地区, 是世界大型食肉动物物种最丰富的地区之一, 而日益增强的人类活动可能会加剧对当地动植物资源的破坏, 进而威胁野生食肉动物的生存。获得准确的物种多样性信息及食肉动物食性数据有助于深入了解该地区生态系统结构及食物网关系, 对研究物种共存机制及生物多样性保护有重要意义。本研究通过从四川甘孜藏族自治州新龙县和石渠县野外采集的食肉动物粪便样品中提取DNA, 利用DNA条形码进行物种鉴定, 快速获得该地区食肉动物物种构成信息。38份粪便样品经鉴定来自于7种食肉动物, 分别为5种大型食肉动物(狼Canis lupus、棕熊Ursus arctos、豹Panthera pardus、雪豹P. unica、狗Canis lupus familiaris)和2种中小型食肉动物(豹猫Prionailurus bengalensis、赤狐Vulpes vulpes)。进一步利用高通量测序和宏条形码技术对7种食肉动物粪便中的食物DNA进行精准食性分析, 得到包含19种哺乳类、8种鸟类和1种鱼类共计28个不同的食物分子可操作分类单元(molecular operational taxonomic unit, MOTU)。结果显示, 狼、狗、棕熊最主要的食物来源为偶蹄目动物, 其中取食频率最高的物种为家牦牛(Bos grunniens); 而豹猫和赤狐食物中小型哺乳动物如啮齿目和兔形目占重要比例, 其中高原松田鼠(Neodon irene)和高原鼠兔(Ochotona curzoniae)被取食频率最高。豹和雪豹的食物分别为偶蹄目的中华斑羚(Naemorhedus griseus)和岩羊(Pseudois nayaur)。本研究显示了粪便DNA及宏条形码技术在食肉动物多样性快速调查及高通量精确食性分析中的应用前景, 并为此类研究提供了技术路线的有力借鉴。
Advances in molecular ecology: Tracking trophic links through predator-prey food-webs
DOI:10.1111/fec.2005.19.issue-5 URL [本文引用: 1]
Spring ashore activity rhythm of Eurasian otters (Lutra lutra) in Changbai Mountain, Jilin Province
吉林长白山地区水獭春季岸上活动节律
Seasonal and habitat-related variation of otter Lutra lutra diet in a Mediterranean river catchment (Italy)
Amphibians in Eurasian otter Lutra lutra diet: Osteological identification unveils hidden prey richness and male-biased predation on anurans
DOI:10.1111/mam.12155
[本文引用: 1]

Amphibians form a major component of the diet of the otter Lutra lutra in several areas of its wide geographic range. Yet, amphibian remains are rarely identified to species level and therefore information on the diversity of this food resource is generally scarce. The aims of this study were: 1) to assess the overall pattern and trends in the use of amphibians as a resource by otters at the range scale, and 2) to highlight current knowledge on the diversity of amphibians taken as prey by otters. Additionally, we carried out osteological identification of amphibian remains in otter spraints (faeces) from southern Italy, with the aim of demonstrating how this method may improve our knowledge on predator-prey relationships. The frequency of occurrence of amphibians in 64 dietary studies averaged 12%. Predation of amphibians by otters increased with longitude and was the highest in the Alpine biogeographical region. Predation by otters was reported on 28 amphibian species (35% of European species). Peaks in their frequency of use were reported for all seasons, mostly in winter and spring. In southern Italy, we identified 355 individuals belonging to at least seven amphibian taxa (64% of available species; Rana italica, Rana dalmatina/italica, Pelophylax kl. bergeri/hispanicus, Hyla intermedia, Bufo bufo, Bufotes balearicus, and Lissotriton italicus), and pointed out male-biased predation within the Order Anura (frogs). We conclude that the contribution of amphibians to the richness of the otter's prey community is far higher than commonly perceived, and that osteological analyses allow the detailed investigation of the feeding behaviour of this top predator of freshwater habitats.
Observer bias and analysis of gray wolf diets from scats
The diet of otters (Lutra lutra L.) in Danish freshwater habitats: Comparisons of prey fish populations
Towards next-generation biodiversity assessment using DNA metabarcoding
DOI:10.1111/j.1365-294X.2012.05470.x
PMID:22486824
[本文引用: 1]

Virtually all empirical ecological studies require species identification during data collection. DNA metabarcoding refers to the automated identification of multiple species from a single bulk sample containing entire organisms or from a single environmental sample containing degraded DNA (soil, water, faeces, etc.). It can be implemented for both modern and ancient environmental samples. The availability of next-generation sequencing platforms and the ecologists' need for high-throughput taxon identification have facilitated the emergence of DNA metabarcoding. The potential power of DNA metabarcoding as it is implemented today is limited mainly by its dependency on PCR and by the considerable investment needed to build comprehensive taxonomic reference libraries. Further developments associated with the impressive progress in DNA sequencing will eliminate the currently required DNA amplification step, and comprehensive taxonomic reference libraries composed of whole organellar genomes and repetitive ribosomal nuclear DNA can be built based on the well-curated DNA extract collections maintained by standardized barcoding initiatives. The near-term future of DNA metabarcoding has an enormous potential to boost data acquisition in biodiversity research.© 2012 Blackwell Publishing Ltd.
The Zoobenthos species in Huma River
呼玛河底栖动物种类组成
Monitoring endangered freshwater biodiversity using environmental DNA
DOI:10.1111/j.1365-294X.2011.05418.x
PMID:22151771
[本文引用: 2]

Freshwater ecosystems are among the most endangered habitats on Earth, with thousands of animal species known to be threatened or already extinct. Reliable monitoring of threatened organisms is crucial for data-driven conservation actions but remains a challenge owing to nonstandardized methods that depend on practical and taxonomic expertise, which is rapidly declining. Here, we show that a diversity of rare and threatened freshwater animals--representing amphibians, fish, mammals, insects and crustaceans--can be detected and quantified based on DNA obtained directly from small water samples of lakes, ponds and streams. We successfully validate our findings in a controlled mesocosm experiment and show that DNA becomes undetectable within 2 weeks after removal of animals, indicating that DNA traces are near contemporary with presence of the species. We further demonstrate that entire faunas of amphibians and fish can be detected by high-throughput sequencing of DNA extracted from pond water. Our findings underpin the ubiquitous nature of DNA traces in the environment and establish environmental DNA as a tool for monitoring rare and threatened species across a wide range of taxonomic groups.© 2011 Blackwell Publishing Ltd.
Fish as predators and prey: DNA-based assessment of their role in food webs
DOI:10.1111/jfb.v98.2 URL [本文引用: 1]
Dietary analysis from fecal samples: How many scats are enough?
DOI:10.1644/1545-1542(2005)086[0704:DAFFSH]2.0.CO;2 URL [本文引用: 1]
Assessing the diet of a predator using a DNA metabarcoding approach
DOI:10.3389/fevo.2022.902412
URL
[本文引用: 7]

The diet of top predators is vital information needed to determine their ecological function and for their conservation management. However, the elusive habit and low population density of many predators constrains determination of their diets. While the morphological identification of scat contents is the traditional method, DNA metabarcoding has lately proven a more efficient and accurate method of identifying prey taxa. We applied DNA metabarcoding to analyzing the diet of the Eurasian otter (Lutra lutra), a top predator in freshwater ecosystems, using 12S and 16S rRNA mitochondrial primers target vertebrate prey. Diet did not vary among different data removal thresholds of 0.1, 1, 3, and 5%, comprising fishes (&gt;90%), amphibians and birds (&gt;2%), and occasionally mammals (&lt;2%). Both 12S and 16S primers revealed similar otter diets, indicating that a single set of primers with a higher threshold is cost-effective for detecting the main prey taxa. Using 12S primers and a 5% threshold, we found no seasonal variation of otter diet in the Tangjiahe National Nature Reserve. A different prey community was found outside the reserve, which resulted in different prey composition for otters. However, prey taxon richness was not different between otters in- and outside the reserve. Otters preferred Schizothorax spp., the largest-sized fish species in the reserve, whereas they mainly preyed on Triplophysa bleekeri, a small-sized fish species, outside the reserve. Otters’ flexible feeding strategy reflect their high adaptability. However, greater human disturbance outside the reserve may present significant challenges to otters by altering prey communities and reducing prey profitability. Combining fecal DNA metabarcoding and local fish survey will provide opportunities for more detailed studies on the impact of different levels of human disturbances on prey communities and otters.
Site-specific and seasonal variation in habitat use of Eurasian otters (Lutra lutra) in Western China: Implications for conservation
Techniques for application of faecal DNA methods to field studies of Ursids
DOI:10.1046/j.1365-294x.1997.00281.x
PMID:9394465
[本文引用: 1]

We describe methods for the preservation, extraction and amplification of DNA from faeces that facilitate field applications of faecal DNA technology. Mitochondrial, protein encoding and microsatellite nuclear DNA extracted and amplified from faeces of Malayan sun bears and North American black bears is shown to be identical to that extracted and amplified from the same individual's tissue or blood. A simple drying agent, silica beads, is shown to be a particularly effective preservative, allowing easy and safe transport of samples from the field. Methods are also developed to eliminate the risk of faecal DNA contamination from hair present in faeces.
Seasonal exploitation of amphibians by otters (Lutra lutra) in north-east Scotland
DOI:10.1111/jzo.1990.220.issue-4 URL [本文引用: 1]
Molecular dietary analysis of two sympatric felids in the Mountains of Southwest China biodiversity hotspot and conservation implications
DOI:10.1038/srep41909
PMID:28195150
[本文引用: 2]

Dietary information is lacking in most of small to mid-sized carnivores due to their elusive predatory behaviour and versatile feeding habits. The leopard cat (LPC; Prionailurus bengalensis) and the Asiatic golden cat (AGC; Catopuma temminckii) are two important yet increasingly endangered carnivore species in the temperate mountain forest ecosystem in Southwest China, a global biodiversity hotspot and a significant reservoir of China's endemic species. We investigated the vertebrate prey of the two sympatric felids using faecal DNA and a next-generation sequencing (NGS)/metabarcoding approach. Forty vertebrate prey taxa were identified from 93 LPC and 10 AGC faecal samples; 37 taxa were found in the LPC diet, and 20 were detected in the AGC diet. Prey included 27 mammalian taxa, 11 birds, one lizard and one fish, with 73% (29/40) of the taxa assigned to the species level. Rodents and pikas were the most dominant LPC prey categories, whereas rodents, pheasant, fowl and ungulates were the main AGC prey. We also analysed the seasonal and altitudinal variations in the LPC diet. Our results provide the most comprehensive dietary data for these felids and valuable information for their conservation planning.
Species of otters in China and the conservation of their natural resources
中国水獭种类及资源保护
Studies on Molecular Phylogeny of Rhynchocypris Fishes (Teleiostei: Cyprinidae) and Phylogeography of Cold-adapting Freshwater Fishes in the Eastern China
鲤科大吻鱥属鱼类的分子系统发育关系与我国东部地区冷水性淡水鱼类的亲缘生物地理
Fish resources in the northern Greater Khingan Mountains
大兴安岭北部鱼类资源现状
Distribution pattern and identification of conservation priority areas of the otter in Northeast China
DOI:10.17520/biods.2021157
[本文引用: 3]

<p id="p00005"><strong>Aims:</strong> The otter is an indicator and flagship species of aquatic ecosystems. Its populations have undergone a drastic decline in China, and have become locally extinct in some regions due to anthropogenic disturbance. However, the current literature on otters in China is inadequate, which has subsequently affected conservation of the species. We aim to assess the potential distribution and conservation priority areas of the Eurasian otter (<i>Lutra lutra</i>) in Northeast China.</p><p id="p00010"><strong>Methods:</strong> In this study, we integrated species distribution models with otter survey data from 2016 to 2020 to assess Eurasian otter distribution. Then, we used the geographic information system and core-area zonation algorithm in Zonation 4.00 to identify conservation priority areas for otter protection. Using the anthropogenic pressures layer, we also evaluated anthropogenic stressors in each province. Then, we analyzed the conservation status of the otter based on the spatial distribution of national nature reserves. Furthermore, we used Inner Mongolia Forest Industry Group, Daxing’anling Forestry Group and Yichun Forest Industry Group as case studies to analyze the role of state-owned forest areas for otter conservation. </p><p id="p00015"><strong>Results:</strong> The results demonstrate that potential distribution and conservation priority areas cover 104,515.04 km<sup>2 </sup>and 45,448.99 km<sup>2</sup>, respectively. Large swathes of conservation priority areas remain for the otter in Daxing’anling which are connected with conservation priority areas in Xiaoxing’anling and there is no obvious geographical boundary between them. Therefore, these regions are mandatory to conserve to maintain the stability of otter populations in Northeast China. Otters in the Liaoning Province faced the greatest anthropogenic pressures, followed by the Jilin Province and Heilongjiang Province. Otters in the Inner Mongolia Autonomous Region faced the lowest amount of pressure. There is potential distribution for the otter in 63 nature reserves and conservation priority areas in 32 nature reserves out of the 110 reserves in the region. The study area includes 11.64% of the otter’s potential distribution and 10.88% of conservation priority areas. Three state-owned forest industry groups cover 71.18% of the potential distribution of the otter (74,390.89 km<sup>2</sup>) and 79.26% of the conservation priority areas (36,022.22 km<sup>2</sup>). </p><p id="p00020"><strong>Conclusions:</strong> This study indicates that state-owned forest areas may play a greater role in the protection of biodiversity following a comprehensive ban on logging in natural forests. Therefore, we propose that areas with important conservation value in state-owned forest land should be gradually included under the protections of national parks in order to achieve systematic and complete protection of biodiversity. Finally, we suggest: (1) strengthening the management of river pollutions; (2) reducing fishing intensity; (3) a long-term field monitoring network be established to study population changes in otters; (4) increasing the investment in scientific research on otters; and (5) raising awareness of otter conservation through public education.</p>
东北地区水獭分布格局与保护优先区识别
DOI:10.17520/biods.2021157
[本文引用: 3]

水獭是水生生态系统重要的指示种和旗舰种, 由于强烈的人为干扰, 中国的水獭种群数量大幅下降, 部分区域已局部灭绝。然而目前国内对水獭的调查和研究非常有限, 本底不清的状况已经严重影响到水獭的野外保育工作。本文以东北地区的欧亚水獭指名亚种(Lutra lutra lutra)为研究对象, 基于2016-2020年的调查数据, 使用组合建模的方法评估了水獭的潜在分布区; 利用地理信息系统和系统保护规划软件分析了水獭的保护优先区并计算了各省级行政区内水獭潜在分布区和保护优先区面临的人类压力; 结合国家级自然保护区的空间布局分析了水獭的保护现状, 并以内蒙古森工集团、大兴安岭林业集团、伊春森工集团三大国有林区为例分析了重点国有林区在水獭保护中的作用。结果表明: (1)水獭潜在分布区和保护优先区面积分别为104,515.04 km<sup>2</sup>和45,448.99 km<sup>2</sup>, 其中大兴安岭的水獭保护优先区集中连片, 并与小兴安岭的保护优先区相连, 栖息地之间没有明显地理隔离, 是维持东北地区水獭种群稳定的重中之重; (2)水獭面临的人类压力大小依次为: 辽宁 > 吉林 > 黑龙江 > 内蒙古; (3)研究区内110个国家级自然保护区中有63个包含水獭潜在分布区, 覆盖面积为12,168.93 km<sup>2</sup>, 仅占水獭潜在分布区面积的11.64%, 其中32个国家级自然保护区包含水獭保护优先区, 占水獭保护优先区面积的10.88%; (4)三大国有林区涵盖了71.18%的水獭潜在分布区和79.26%的保护优先区(面积分别为74,390.89 km<sup>2</sup>和36,022.22 km<sup>2</sup>)。由此可见, 尽管水獭潜在分布区中国家级自然保护区占比较低, 但是在天然林全面禁伐的背景下, 重点国有林区可能在未来东北地区的生物多样性保护中发挥更大作用, 因此我们建议将重点国有林区中具有重要保护价值的区域逐步纳入以国家公园为主体的自然保护地体系中, 以实现生物多样性的系统性和完整性保护。最后, 本文结合研究结果和实地调研提出以下保护建议: (1)加强对河流污染物的管理; (2)控制渔民捕鱼强度; (3)开展全面的水獭专项调查并建立长期的监测体系; (4)加大对水獭的科研投入; (5)加强宣传力度, 提升公众保护意识。
The neglected otters in China: Distribution change in the past 400 years and current conservation status
DOI:10.1016/j.biocon.2018.10.028 URL [本文引用: 2]