物种共存机制是生态学的核心课题之一, 揭示物种共存的发生和维持机制对于群落生态学理论发展具有重要意义, 也是生物多样性研究的热点之一。生态学家提出了多种揭示共存机制的理论和假说, 其中包括资源比例假说(Huisman & Weissing, 1999)、中度干扰假说(Sommer, 1999)和生态位分化理论(Grinnell, 1917; Udvardy, 1959)。目前, 针对野生动物同域共存的研究大多集中在鸟类(De León et al, 2014; Hou et al, 2021)、灵长类(Porter, 2001; 王丞等, 2020)、食草动物(Liu et al, 2007; Kartzinel et al, 2015)等大中型动物, 而对物种数量多、分布范围广的小型哺乳动物同域共存机制研究较为匮乏。
生存需求相似的物种往往伴随着生态位的分化(Hardin, 1960), 竞争的结果将导致食物、栖息地和时间等资源的分化, 进而促进生境内多物种共存。国内外学者围绕生态位分化理论的研究主要从时间生态位(Rutrough et al, 2019)、空间生态位(杨春文, 2008)和营养生态位(Cooke & Crowley, 2018)等方面展开。其中, 食性分析是营养生态位研究的重要组成部分, 是动物根据自身营养需求对生境适应的结果(Marshal et al, 2008), 营养生态位即动物对食物资源的利用情况。食性分析是一项重要的基础研究内容(Cooke & Crowley, 2018), 可以直观展现动物的营养需求特征, 反映出动物的生态功能及其营养关系(殷宝法等, 2007)。食物也为动物生长发育和生存繁衍提供必需的营养成分, 是探讨食物网营养流动、评估物种生存状况及种间关系等热点问题的重要基础(钟林强, 2020; 张宇洋等, 2022)。
小型哺乳动物作为生态系统的消费者, 在能量流动和物质循环等过程中至关重要, 并通过觅食、挖掘巢穴等活动影响生态系统的结构和功能(雍仲禹等, 2011)。因此, 该类动物对维持生态平衡具有重要意义。但在其种群数量激增时也会对生态系统造成严重危害, 损害农业、林业、畜牧业经济(王德利等, 2016)。森林中的小型哺乳动物会取食树木种子、剥食树皮和嫩芽, 部分甚至食用树根, 对树木乃至森林造成严重危害(王贵来, 2004)。多数小型哺乳动物也是各种疾病的传染源, 携带多种病原菌, 在公共卫生领域也被高度重视(邓址, 1991)。因此, 明确其食物组成和摄食策略有助于从营养生态位层面揭示多种小型哺乳动物同域共存机制, 也为小型哺乳动物物种多样性的维持和种群生态管理提供理论依据。
动物食性研究方法众多, 如直接观察法(多用于观察取食和贮食的行为特征)、实验室笼养饲喂法(更适用于研究动物对食物的偏好选择)、颊囊内含物分析法(只适用于有颊囊的动物, 研究对象受限)、稳定同位素技术(难以确定具体食物组成)、胃内容物显微组织分析法、粪便显微分析法以及DNA宏条形码技术等(陆琪等, 2019; Shutt et al, 2020; Shao et al, 2021; 赵耕, 2021)。基于粪便对食物进行传统形态学鉴定也有诸多问题: (1)对外观相似的食物无法准确区分; (2)肌肉、软组织等部分食物消化完全, 难以鉴定; (3)要求实施者具备高水平的分类学技能; (4)总体鉴定效率低下, 准确率有待提升等(Pompanon et al, 2012; Akrim et al, 2018)。目前, DNA宏条形码技术在动物食性研究领域潜力巨大, 极大地提高了食性分析的精确性(Taberlet et al, 2012)。与传统方法相比, DNA宏条形码技术优势明显: (1)几乎不受食物消化影响, 可鉴定出形态上无法识别的无脊椎动物; (2)提高了物种鉴定的准确度, 耗时少; (3)不依靠分类学经验; (4)可同时分析多个样本(Razgour et al, 2011; Pompanon et al, 2012; Yang et al, 2016)。该技术已广泛应用于食草动物(Kartzinel et al, 2015; 郭艳萍等, 2021)、食肉动物(陆琪等, 2019; 邵昕宁等, 2019)和杂食动物(Tang et al, 2020; Hou et al, 2021)的分子食性研究。
四川老君山国家级自然保护区内分布着多种小型哺乳动物, 通过我们前期调查, 其中以4种啮齿目动物巢鼠(Micromys minutus)、社鼠(Niviventer confucianus)、针毛鼠(N. fulvescens)、黄胸鼠(Rattus tanezumi)及2种劳亚食虫目动物四川短尾鼩(Anourosorex squamipes)和淡灰黑齿鼩鼱(Blarinella griselda)居多。它们的生存环境与生活习性等方面存在诸多相似, 是研究小型哺乳动物同域共存机制的理想对象。本研究拟基于DNA宏条形码技术, 对6种小型哺乳动物的动植物食物组成进行鉴定和比较分析, 并计算种间食物生态位重叠指数和食物生态位宽度, 从营养生态位层面探究该保护区内多种小型哺乳动物同域共存的机制。
1 研究方法
1.1 研究区域
四川老君山国家级自然保护区(103º57'- 104º04' E, 28º39'-28º43' N)地处四川省宜宾市屏山县, 保护区位于凉山山系, 区内海拔900-2,008 m, 垂直气候差异大, 东西约为11.6 km, 南北约为7.4 km, 总面积为35 km2, 是以保护四川山鹧鸪(Arborophila rufipectus)等雉科鸟类为主的自然保护区, 是全球生物多样性保护的热点地区, 其他野生动物资源也十分丰富(付义强等, 2016)。保护区为亚热带湿润型季风气候, 雨量充沛, 年平均气温12-14℃, 气候湿润, 适宜小型哺乳动物生存。阔叶林为该生态系统的主要植被类型, 为多种野生动物提供了适宜的栖息地和觅食场所(Liao et al, 2007, 2008; 西丽媛子等, 2020)。
1.2 样本采集
本研究于2020年夏季在该保护区核心区的3个不同海拔段采用铗夜法和陷阱法对目标物种进行捕获, 每个海拔段采集6种小型哺乳动物各3只。所有采集的动物标本均在现场根据外部特征进行初步形态学鉴定。每只标本取少量肝脏或肌肉组织保存于酒精中, 于实验室-40℃冰柜保存, 以备后续DNA的提取。在胃内容物食糜DNA提取前, 混合同一海拔采集的同一物种的3份胃内容物样本。最终参与测序的样品为每个物种在3个不同海拔段的3份食糜DNA, 因此共18份胃内容物食糜DNA被用于后续测序。在保护区管理站对动物标本进行解剖, 使用一次性采样拭子收集其肠、胃内容物保存于装有常温细胞保存液的冻存管中, 用冰袋冷藏运输, 送回实验室后置于-40℃冰柜保存。目标物种的分子鉴定按如下步骤进行: (1)提取收集的肝脏或肌肉组织的总DNA; (2) PCR扩增; (3) PCR产物纯化回收和Sanger测序; (4)序列BLAST比对和物种鉴定。组织样品总DNA提取使用动物组织DNA提取试剂盒(成都福际生物公司), 选择引物对L14724-hk3和H15915-hk3 (He et al, 2010) (附录1)扩增Cyt b基因序列。PCR目的产物通过凝胶电泳成像, 将符合目的片段大小的PCR产物纯化回收后由擎科生物科技有限公司(成都)进行双向测序。测序结果在NCBI (
1.3 食物DNA扩增和测序
胃内容物中食糜DNA的提取采用QIAamp DNA Stool Mini Kit试剂盒。由于胃内容物中的食物DNA存在降解和PCR抑制等问题, 因此较短的扩增片段可能会提高扩增成功率(陆琪等, 2019)。本研究分别选用线粒体COI (cytochrome c oxidase subunit I, 线粒体细胞色素C氧化酶亚基)和RbcL (ribulose- bisphosphate carboxy lase gene, 核酮糖-1,5-二磷酸羧化酶基因)鉴定动植物性食物(Hofreiter et al, 2000), 引物分别为COIZBJ-ArtF1c/COIZBJ-ArtR2c (Burgar et al, 2014; Clare et al, 2014)和Z1aF/hp2R (附录1), 连接的样本特异性barcode序列见附录2。PCR扩增体系为20 μL, COI引物PCR扩增程序为98℃预变性5 min; 95℃变性30 s, 53℃退火30 s, 68℃延伸30 s, 进行5个循环; 98℃变性30 s, 45℃退火30 s, 68℃延伸45 s, 进行30个循环; 68℃延伸5 min, 在12℃下保存。RbcL引物PCR扩增程序为98℃预变性5 min; 98℃变性30 s, 52℃退火30 s, 72℃延伸45 s, 进行30个循环; 72℃延伸5 min, 12℃保存。PCR扩增使用全式金公司的Pfu高保真DNA聚合酶。每个PCR板和不同引物组设置阴性对照用于检测环境、试剂等污染。采用DNA纯化磁珠(Vazyme VAHTSTM DNA Clean Beads)对PCR扩增产物进行纯化回收。以纯化后的PCR产物为模板, 采用荧光试剂(Quant-iT PicoGreen dsDNA Assay Kit)进行荧光定量。根据高通量测序要求, 将每个样品的扩增子按比例混合和纯化回收, 使用Illumina公司的TruSeq Nano DNA LT Library Prep Kit制备测序文库, 文库的高通量双末端测序在上海派森诺生物信息技术公司的Illumina NovaSeq 6000测序平台进行, 每端读长为250 bp。
1.4 序列处理与数据统计分析
经高通量测序获得的原始数据需通过修剪和质量控制步骤进行筛选(Caporaso et al, 2012)。使用Vsearch软件对扩增子序列完成合并、质量过滤、去重、去嵌合体、聚类等步骤(Edgar, 2013)。基于操作分类单元(operational taxonomic unit, OTU)聚类的Vsearch (Rognes et al, 2016)方法, 对有效序列进行OTU归并划分, 以98%的序列相似度作为OTU划分的阈值。在NCBI数据库中使用BLAST比对通过质量过滤的序列(Berry et al, 2017), 根据相似度百分比阈值, 序列被鉴定到种、属等分类单位。序列比对结果一致性不低于99%, 且最匹配序列只对应单一物种, 同时该物种在当地有分布时, 认为该序列来自于该物种; 序列比对一致性不低于98%被认为是属水平的注释。本研究的原始测序数据已上传至组学原始数据归档库(genome sequence archive, GSA), 动物性食物数据序列号为CRA007014; 植物性食物数据序列号为CRA007015。
α多样性的计算与分析在QIIME2软件中完成。为了评估测序深度的充分性, 本研究根据OTU的数量绘制了样本的稀疏曲线。基于Jaccard距离矩阵进行了非量度多维尺度分析(nonmetric multidimensional scaling analysis, NMDS), 并使用R v.3.3.3的ggplot2包(Wickham, 2009)进行图形可视化分析, 且使用ANOSIM (analysis of similarities)进行组间差异显著性分析。本研究使用相对丰度(relative abundance, RA)和出现频率(frequency of occurrence, FO)来衡量食物组成和出现情况(Deagle et al, 2019)。使用LEfSe (linear discriminatory analysis (LDA) effect size)分析, 在不同分类水平上鉴定了劳亚食虫类四物种和啮齿目两物种的主要差异性食物。为比较食物组成和多样性在不同物种中的显著性差异情况, 本研究使用SPSS 20的单因素方差分析(ANOVA)和Student’s t检验检测差异显著性。
1.5 营养生态位宽度和重叠指数
式中, B代表生态位宽度, Pj代表食物j所占的比例;
式中, Pij和Pik分别代表食物i在物种j和k的食物中所占的相对丰度。Ojk的变化范围为0-1, 根据Krebs (1999)的评判标准, Ojk > 0.3视为重叠有意义, Ojk > 0.6视为显著重叠。
2 结果
2.1 6种小型哺乳动物的食物组成
基于COI宏条形码的测序数据, 所有物种的胃内容物样品共获得2,029,373条有效序列, 平均每个样品112,743 ± 41,572条。随着测序深度的增加, 样品稀疏曲线逐渐达到平稳状态, 表明本研究测序深度合理(附录3)。
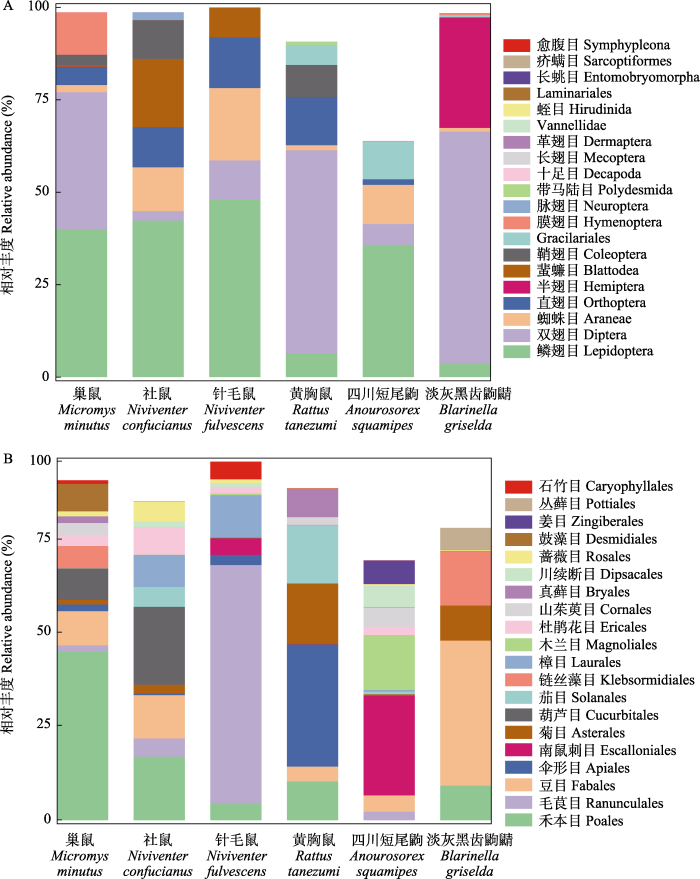
6种小型哺乳动物的动物性食物主要由无脊椎动物构成, 其中在门水平占比较多的为节肢动物门, 其相对丰度(RA)占所有动物性食物的88.31%; 纲水平占比较多的食物为昆虫纲(RA = 79.02%)。在目水平, 鳞翅目、双翅目和蜘蛛目构成这6种小型哺乳动物的主要动物性食物(图1A), 这3个目总共占全部动物性食物的62.10%。其中在巢鼠、社鼠、针毛鼠和四川短尾鼩4个物种的胃内容物中检测到大量鳞翅目昆虫, 其相对丰度分别为39.87%、42.33%、47.98%、35.76% (附录4)。而双翅目的序列在巢鼠(37.19%)、黄胸鼠(55.02%)和淡灰黑齿鼩鼱(62.63%)的食物组成中相对丰度较高。在淡灰黑齿鼩鼱的食物中检测到较多的半翅目(29.69%)昆虫, 而其余5种小型哺乳动物食物中的半翅目相对丰度则较低(0-0.05%), 直翅目的相对丰度较高(1.48%- 13.60%)。在属水平, 黄胸鼠和淡灰黑齿鼩鼱食物中壳粗腹摇蚊属(Conchapelopia, 隶属于双翅目)的相对丰度较高(分别为34.83%、30.52%) (附录5); 美舟蛾属(Uropyia, 隶属于鳞翅目)动物构成了社鼠(16.47%)、针毛鼠(18.08%)和四川短尾鼩(30.35%)动物性食物的主要部分。巢鼠大量摄食长喙天蛾属(Macroglossum, 隶属于鳞翅目, 29.40%)和尺禾蝇属(Geomyza, 隶属于双翅目, 27.72%)物种, 而短痣蚜属(Anoecia, 隶属于同翅目, 29.68%)在淡灰黑齿鼩鼱的食物中被大量检测到。
图1
图1
6种小型哺乳动物目水平排名前20的动物性食物(A)和植物性食物(B)的相对丰度
Fig. 1
Relative abundance of top 20 animal- (A) and plant-derived (B) food items at the order level in the six small mammals
基于RbcL宏条形码鉴定植物性食物的数据中, 所有样本高通量测序后共获得1,581,373条有效序列, 平均每个样品87,854 ± 37,783条。在纲水平, 6种小型哺乳动物植物性食物占比较多的为百合纲植物, 相对丰度为19.21%。在目和属水平, 鉴定到的分类单元相对丰度超过1%的分别有20个目和16个属。其中, 禾本目和豆目构成6种小型哺乳动物的主要植物性食物来源, 伞形目也表现出较高的相对丰度(图1B), 这3个目共占植物性食物的37.20%。除四川短尾鼩大量取食南鼠刺目(26.71%)食物外, 其余5种小型哺乳动物取食较多的为禾本目植物, 其中禾本目序列在巢鼠植物性食物中占比最高(45.33%)。针毛鼠取食毛茛目(63.74%)的比例最高, 其余5种小型哺乳动物则不喜取食毛茛目食物, 而对豆目植物较为青睐(附录6)。相比于其他物种, 黄胸鼠食物中伞形目的相对丰度较高(32.81%), 葫芦目(20.90%)在社鼠食物中的比例较高, 菊目植物则是巢鼠、黄胸鼠和淡灰黑齿鼩鼱都较为喜好的食物。在属水平的植物性食物中, 八月瓜属(Holboellia, 隶属于毛茛目)在针毛鼠食物中较为丰富(60.89%) (附录7)。巢鼠、黄胸鼠和社鼠分别喜好取食芒属(Miscanthus, 隶属于禾本目, 33.37%)、当归属(Angelica, 隶属于伞形目, 32.69%)和绞股蓝属(Gynostemma, 隶属于堇菜目, 20.18%)植物。燕麦属(Avena, 隶属于禾本目)则是巢鼠、社鼠、黄胸鼠和淡灰黑齿鼩鼱都青睐的食物。以上结果与LEfSe分析结果相符合(附录8), 表明在不同分类水平下, 6种小型哺乳动物主要食物的摄食比例存在差异, 同时某些植物(如菊目和燕麦属植物)是它们共同的食物。
2.2 食物组成多样性的种间比较
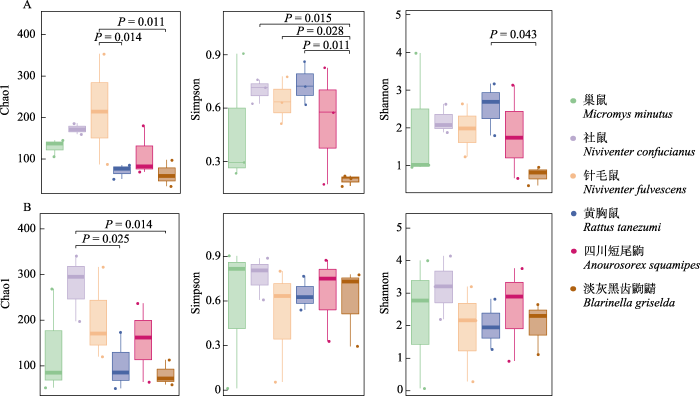
α多样性指数(Chao1、Shannon和Simpson)计算结果表明, 6种小型哺乳动物的动物性食物多样性存在一定的种间差异(图2A)。针毛鼠的Chao1丰富度指数显著高于淡灰黑齿鼩鼱(P = 0.011), 黄胸鼠和针毛鼠的Chao1丰富度指数差异显著(P = 0.014)。淡灰黑齿鼩鼱的Simpson指数显著低于社鼠(P = 0.015), 同时显著低于针毛鼠(P = 0.028)和黄胸鼠(P = 0.011)。淡灰黑齿鼩鼱的Shannon指数显著低于黄胸鼠(P = 0.043)。3个α多样性指数的种间差异比较显示, 淡灰黑齿鼩鼱的动物性食物多样性水平最低。总体上, 植物性食物的α多样性在6种小型哺乳动物之间差异并不显著(图2B)。只有社鼠的Chao1丰富度指数显著高于淡灰黑齿鼩鼱(P = 0.014), 黄胸鼠与社鼠的Chao1丰富度指数差异显著(P = 0.025)。
图2
图2
6种小型哺乳动物动物性食物(A)和植物性食物(B)的α多样性指数的分组箱线图
Fig. 2
Box-and-whisker plots for α diversity of animal- (A) and plant-derived (B) food items of six small mammals
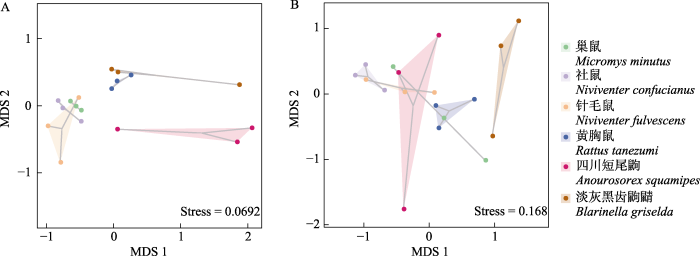
基于Jaccard distance矩阵的NMDS聚类结果显示了6种小型哺乳动物的动植物食物组成的种间差异情况(图3)。四川短尾鼩的动物性食物组成与其他5种小型哺乳动物明显分离(ANOSIM: R = 0.259- 0.778, P = 0.092-0.302)。巢鼠、社鼠和针毛鼠的动物性食物相对聚集, 且有一定重叠。黄胸鼠和淡灰黑齿鼩鼱的动物性食物聚集距离较近。此外, 淡灰黑齿鼩鼱的植物性食物与其他5种小型哺乳动物存在一定分离(ANOSIM: R = -0.111至0.519, P = 0.101-0.689)。四川短尾鼩和巢鼠、针毛鼠的植物性食物的重叠较多。
2.3 食物生态位宽度和食物重叠
在6种小型哺乳动物的属水平营养生态位宽度计算结果中, 社鼠的动物性食物营养生态位宽度最广(B = 8.6485), 其次是针毛鼠(B = 8.1913), 四川短尾鼩(B = 4.0286)和淡灰黑齿鼩鼱(B = 3.7137)的较低(表1)。本研究中的4种啮齿目物种的动物性食物平均营养生态位宽度均显著高于2种劳亚食虫目物种(B: 7.1181 v.s. 3.8712, P = 0.003)。在植物性食物营养生态位宽度计算结果中, 社鼠的营养生态位宽度最大(B = 11.0998), 其后依次为巢鼠(B = 6.9642)、四川短尾鼩(B = 5.8556)、黄胸鼠(B = 5.5751)、淡灰黑齿鼩鼱(B = 4.5893), 针毛鼠的最低(B = 2.5601)。总体上, 6种小型哺乳动物的植物性食物营养生态位宽度差异较大。4种啮齿目物种的植物性食物平均营养生态位宽度与2种劳亚食虫目物种间并无显著差异。同时, 科水平的营养生态位宽度计算结果与属水平的结果较为一致(表1)。
表1 6种小型哺乳动物的营养生态位宽度
Table 1
| 动物性食物 Animal-derived food | 植物性食物 Plant-derived food | |||
|---|---|---|---|---|
| 属水平 Genus level | 科水平 Family level | 属水平 Genus level | 科水平 Family level | |
| 巢鼠 Micromys minutus | 5.4882 | 5.4409 | 6.9642 | 4.7517 |
| 社鼠 Niviventer confucianus | 8.6485 | 8.2191 | 11.0998 | 8.6986 |
| 针毛鼠 Niviventer fulvescens | 8.1913 | 6.4274 | 2.5601 | 2.5435 |
| 黄胸鼠 Rattus tanezumi | 6.1445 | 6.1359 | 5.5751 | 5.5339 |
| 四川短尾鼩 Anourosorex squamipes | 4.0286 | 4.0239 | 5.8556 | 5.8525 |
| 淡灰黑齿鼩鼱 Blarinella griselda | 3.7137 | 3.7118 | 4.5893 | 4.5681 |
在6种小型哺乳动物的属水平营养生态位重叠指数计算结果中, 黄胸鼠和淡灰黑齿鼩鼱的动物性营养生态位重叠指数最高(Ojk = 0.63), 表明二者营养生态位重叠明显。社鼠与针毛鼠(Ojk = 0.31)、社鼠与四川短尾鼩(Ojk = 0.40)、针毛鼠与四川短尾鼩(Ojk = 0.35)的动物性食物有一定程度重叠(表2), 表明它们的生活史中可能面临动物性食物资源的竞争。其余几种小型哺乳动物两两间的动物性食物重叠指数均较低(表2)。在植物性营养生态位重叠指数中, 社鼠和淡灰黑齿鼩鼱的食物生态位重叠指数最高(Ojk = 0.48), 巢鼠和社鼠(Ojk = 0.34)、四川短尾鼩和淡灰黑齿鼩鼱(Ojk = 0.31)也表现出一定程度的食物生态位重叠, 其余小型哺乳动物两两间的食物生态位重叠指数均较低。此外, 本研究还计算了4种啮齿目物种间的营养生态位重叠指数, Ojk值均小于0.01。
表2 6种小型哺乳动物属水平的动物性(斜线左)和植物性(斜线右)食物营养生态位重叠指数
Table 2
| 巢鼠 Micromys minutus | 社鼠 Niviventer confucianus | 针毛鼠 Niviventer fulvescens | 黄胸鼠 Rattus tanezumi | 四川短尾鼩 Anourosorex squamipes | |
|---|---|---|---|---|---|
| 社鼠 Niviventer confucianus | 0.04/0.34 | ||||
| 针毛鼠 Niviventer fulvescens | 0.07/0.09 | 0.31/0.22 | |||
| 黄胸鼠 Rattus tanezumi | 0.08/0.12 | 0.08/0.24 | 0.14/0.01 | ||
| 四川短尾鼩 Anourosorex squamipes | 0.01/0.06 | 0.40/0.26 | 0.35/0.12 | 0.21/0.12 | |
| 淡灰黑齿鼩鼱 Blarinella griselda | 0.04/0.23 | 0.01/0.48 | 0.02/0.01 | 0.63/0.24 | 0.03/0.31 |
根据Krebs (
According to Krebs (
3 讨论
3.1 食物组成特点
本研究基于DNA宏条形码技术解析了老君山国家级自然保护区6种优势小型哺乳动物夏季的食物组成和食物资源种间竞争关系。结果表明, 鳞翅目、双翅目昆虫和蜘蛛目是6种小型哺乳动物主要的动物性食物, 禾本目和豆目构成了其大部分植物性食物。不同小型哺乳动物之间虽然存在食物生态位重叠, 但在主要食物上出现分离, 表明它们可能通过摄食不同比例和种类的食物来降低种间食物资源竞争, 以实现同域共存。
本研究较之前关于同域共存小型哺乳动物营养生态位的报道(李天保, 2020; 张彩军等, 2021), 获得了分辨率更高的动植物食物图谱。同域分布物种营养生态位上的分化在其他同域共存小型哺乳动物研究中也有报道。如, 陈文文等(2014)通过对神农架地区4种啮齿动物的食物组成进行比较, 发现安氏白腹鼠(Niviventer andersoni)、社鼠、中华姬鼠(Apodemus draco)和高山姬鼠(A. chevrieri)的摄食偏好有所差异。安氏白腹鼠主要食用植物枝叶和种子, 其余3种鼠类则主要食用种子和动物性食物。本研究中, 社鼠、针毛鼠和四川短尾鼩均取食较多的美舟蛾属食物, 黄胸鼠和淡灰黑齿鼩鼱取食壳粗腹摇蚊属动物较多(附录5)。主要食物组成相对丰度差异(图1)和食物聚类分析结果(图3)显示, 6种小型哺乳动物在主要食物的摄食偏好性上存在一定差异, 表明它们可以通过不同取食策略达到摄食比例的差异, 以减少对栖息地食物资源的竞争, 实现长期共存。此外, 了解小型哺乳动物的食物偏好性, 对于其防治也有重要意义。王明春等(1997)对甘肃鼢鼠(Eospalax cansus)食物偏好性的研究发现, 玉米是其最喜食的饵料, 在防治甘肃鼢鼠时, 可以使用玉米作饵料诱杀。李新民等(1989)发现褐家鼠(Rattus norvegicus)、黄胸鼠对大米的偏好性要优于小麦和玉米, 但对炒米的喜食性降低, 不宜用炒米配置毒饵。本研究结果表明, 6种小型哺乳动物在动植物食物中各有其偏好性食物, 在鼠形动物的防治管理中可提供参考依据。
图3
图3
基于Jaccard distance矩阵的6种小型哺乳动物的动物性食物(A)和植物性食物(B)的NMDS图
Fig. 3
The NMDS of animal- (A) and plant-derived (B) food items of six small mammals based on Jaccard distance
本研究中, 四川短尾鼩的动物性食物主要由鳞翅目、双翅目昆虫和蜘蛛目构成, 该结果与其他鼩鼱属动物的食物组成相似。例如, 在南短尾鼩鼱(Blarina carolinensis)的食物谱组成中, 昆虫是其主要食物来源, 特别是双翅目和膜翅目动物, 共约占其食物的23% (Hoffman et al, 2012)。此外一定比例的蜘蛛目、鞘翅目、端足目和腹足纲等无脊椎动物也在南短尾鼩鼱的胃内容物中被检测到(Calhoun, 1941; Jow et al, 1994; Whitaker, 2006; Ladine & Munoz, 2010)。在水鼩鼱(Neomys fodiens)的粪便颗粒中鉴定出最常见的食物为单向蚓目、等足目和双翅目(Churchfield, 1979)。Tang等(2021)通过对四川短尾鼩四季食性的研究, 发现四川短尾鼩全年动物性食物主要为单向蚓目动物, 本文与前人存在差异, 推测可能与采样地的生境类型(成都平原和四川老君山国家级自然保护区)和食物资源空间分布差异有关。在植物性食物中, 以豆目和禾本目食物较多(Tang et al, 2021), 本研究与其一致。另外, 小型哺乳动物的捕食对象和效率也会受到昆虫生活史特征变化(例如, 发育阶段、体型)的影响(Sullins et al, 2018), 这也可能造成不同研究中的优势食物组成结果存在差异。另外, 本研究取样时间单一只能反映6种小型哺乳动物夏季的食物组成特征, 可能造成对其食物多样性的低估。
3.2 营养生态位宽度与重叠
生态位宽度表示生物对资源的利用和对生境的适应能力(Crozier, 1985), 营养生态位宽度代表动物在生态环境中的营养层及营养关系。最优觅食理论预测, 在食物丰富度高的季节, 营养生态位宽度会降低, 而在食物丰富度低的季节, 营养生态位宽度会增加(MacArthur & Pianka, 1966)。本研究中, 社鼠的动物和植物食物生态位宽度指数均最高(表1), 表明社鼠对食物资源的利用能力和对生境的适应能力相对较强。针毛鼠的动物性食物营养生态位宽度较宽, 但植物性食物营养生态位宽度最窄(表1), 暗示其摄食动物性食物的多样性较高。在针毛鼠植物性食物中检测出八月瓜属食物的含量最多(RA = 60.9%), 可能是造成其植物性食物α多样性较低的原因之一。老君山地区八月瓜属植物资源丰富, 尤其在果期可充分满足针毛鼠食物需求, 表明针毛鼠对八月瓜属的摄食偏好性很高。
生态位重叠指数反映了物种间对生境内资源的利用程度, 并在一定程度上衡量物种间潜在的竞争关系(王桂明等, 1996)。营养生态位重叠较高的物种, 可通过改变觅食场所、调整活动节律、取食不同类型的食物等方式以减轻种间竞争强度。营养生态位重叠指数也能反映物种间食性的相似程度。Biffi等(2017)发现比利牛斯鼬鼹(Galemys pyrenaicus)和水鼩鼱通过改变取食行为或觅食微生境差异来实现对营养资源的差异利用, 以实现二者的共存。本研究采样在夏季进行, 研究地在该季节植物性食物多样性较高, 可能会增加捕食者之间的食物重叠。如黄胸鼠和淡灰黑齿鼩鼱的动物性食物存在显著重叠(Ojk = 0.63), 但在食物资源充盈的季节, 明显的食物重叠可能并不会造成种间食物竞争的加剧。成功的资源分配也可能与捕食者如何根据其觅食策略获取食物资源有关。根据觅食模式和觅食微栖息地的差异, 营养生态位的分离现象在鼩鼱中被报道(Churchfield & Rychlik, 2006)。Churchfield和Rychlik (2006)认为, 体型在资源分配中起重要作用, 可导致不同的觅食模式。由于体型大小的差异(如本研究中四川短尾鼩和淡灰黑齿鼩鼱的头体长分别为74-110 mm和65-81 mm), 同域物种可以在同一栖息地捕食不同发育阶段和体型大小的猎物。
食物分化通常被理解为物种相对于主要食物类型的专一化(Churchfield & Sheftel, 1994)。本研究中, 黄胸鼠和淡灰黑齿鼩鼱均取食壳粗腹摇蚊属物种, 其动物性食物营养生态位显著重叠(Ojk = 0.63), 但淡灰黑齿鼩鼱的食物中短痣蚜属(Anoecia)的摄入比例显著高于黄胸鼠(29.7% v.s. 0.001%, P < 0.01), 表明淡灰黑齿鼩鼱可能通过摄食更多的半翅目昆虫以减少同黄胸鼠的食物竞争。其余发生显著重叠的不同物种之间, 均发现两两比较的物种会取食同一属食物, 但在各自的专性食物的摄食比例上又彼此分开。此外, 动物性食物出现重叠的物种间在植物性食物中的重叠指数都较低(表2)。因此, 该地区6种小型哺乳动物夏季可能通过改变主要食物的摄食比例以减少营养生态位的重叠, 避免摄食冲突。
与传统方法相比, DNA宏条形码技术虽然实现了高分辨率的食物鉴定, 但也存在不足。由于动物的二级进食, 使用分子工具可能鉴定出额外的食物分类单元(Clare et al, 2014), 因此基于分子手段获得的食物数据往往低估了常见主要食物种类的重要性, 而可能高估稀少食物(Krüger et al, 2014)。本研究在属水平鉴定出多种相对丰度极低的动植物类群, 其中一部分食物可能为二级进食导致。如很多昆虫可取食植物, 植食性食物中较多低丰度的食物可能是由小型哺乳动物取食昆虫带入。由于DNA宏条形码技术基于食糜中的DNA序列来获取食物组成情况, 测序结果也会受到多种因素的影响, 如标记基因、基因区域和引物的选择, 特异性阻断引物的扩增成功率以及DNA条形码参考数据库的完善程度等(王雪芹等, 2017; Jusino et al, 2019)。因此, 采用多种食性研究方法是补充数据和验证结果准确性的有效途径(刘刚等, 2018)。小型哺乳动物在研究地分布广泛, 但本研究涉及的动物标本数量较少, 取样范围较窄, 可能会对其食物组成的表征不够全面, 从而影响对物种间食物资源利用偏好的解读。此外, 由于本研究取样时间单一, 即使在6种小型哺乳动物的主要食物存在组成上的差异, 也不能推测物种间的营养生态位分化在全年内都存在。取样的时间尺度也是影响判断物种间生态位是否分化的因素之一。在未来的研究中, 基于更大的样品量, 更长时间跨度和地理尺度研究同域物种共存机制可能更有价值。
基于DNA宏条形码技术, 本研究对四川老君山国家级自然保护区优势分布的6种小型哺乳动物开展分子食性研究。通过鉴定食物种类及明确取食偏好性进一步了解小型哺乳动物间的营养生态位分化和重叠程度及潜在的种间竞争关系。该自然保护区分布的6种小型哺乳动物作为生态系统的消费者, 对于维持老君山生态系统的能量流动和物质循环等方面具有重要意义。对6种小型哺乳动物食物组成及营养生态位的研究, 为从营养生态位分化角度揭示物种同域共存机制提供理论依据。此外, 研究结果对了解老君山国家级自然保护区小型哺乳动物的多样性及其维持机制具有借鉴意义, 并将为该地区鼠形动物种群管理提供理论支持。
附录 Supplementary Material
附录1 本研究使用的引物序列
Appendix 1 Sequences of the primers used in the study
附录2 本研究使用的各样品对应的barcode序列
Appendix 2 Barcode sequences of the samples used in the study
附录3 6种小型哺乳动物基于样本的稀疏曲线
Appendix 3 Sample-based rarefaction curves of the six small mammals
附录4 6种小型哺乳动物目水平食物中排名前10的动物性食物的出现频率(FO)和平均相对丰度(RA)
Appendix 4 The frequency of occurrence (FO) and average relative abundance ± standard error (RA ± SE) of the top 10 animal food items at the order level in the diet of the six small mammals
附录5 6种小型哺乳动物属水平食物中排名前10的动物性食物的出现频率(FO)和平均相对丰度(RA)
Appendix 5 The frequency of occurrence (FO) and average relative abundance ± standard error (RA ± SE) of the top 10 animal food items at the genus level in the diet of the six small mammals
附录6 6种小型哺乳动物目水平食物中排名前10的植物性食物的出现频率(FO)和平均相对丰度(RA)
Appendix 6 The frequency of occurrence (FO) and average relative abundance ± standard error (RA ± SE) of the top 10 plant food items at the order level in the diet of the six small mammals
附录7 6种小型哺乳动物属水平食物中排名前10的植物性食物的出现频率(FO)和平均相对丰度(RA)
Appendix 7 The frequency of occurrence (FO) and average relative abundance (RA) ± standard error (RA ± SE) of the top 10 plant food items at the genus level in the diet of the six small mammals
附录8 6种小型哺乳动物的动物性和植物性食物的LEfSe分析结果
Appendix 8 The result of LEfSe analysis of animal- and plant-derived food items in six small mammals
致谢
本研究在样品的野外采集过程中得到了四川大学李凤君博士研究生和四川省林业科学研究院的大力支持, 在此一并感谢。
参考文献
Assessment of bias in morphological identification of carnivore scats confirmed with molecular scatology in north-eastern Himalayan region of Pakistan
DOI:10.7717/peerj.5262
URL
[本文引用: 1]

Scats are often used to study ecological parameters of carnivore species. However, field identification of carnivore scats, based on their morphological characteristics, becomes difficult if many carnivore species are distributed in the same area. We assessed error rates in morphological identification of five sympatric carnivores’ scats in north-eastern Himalayan region of Pakistan during 2013–2017. A sample of 149 scats were subjected to molecular identification using fecal DNA. We used a confusion matrix to assess different types of errors associated with carnivore scat identification. We were able to amplify DNA from 96.6% (n = 144) of scats. Based on field identification of carnivore scats, we had predicted that out of 144 scats: 11 (7.6%) scats were from common leopard, 38 (26.4%) from red fox, 29 (20.1%) from Asiatic jackal, 37 (25.7%) from yellow throated martin, 14 (9.7%) from Asian palm civet and 15 (10.4%) from small Indian civet. However, molecular identification revealed and confirmed nine were scats (6.24%) from common leopard, 40 (27.8 %) from red fox, 21 (14.6%) from Asiatic jackal, 45 (31.25%) from Asian palm civet, 12 (8.3%) scats from small Indian civet, while 11 scats (7.6%) were found from Canis lupus Spp., three (2%) from dog, one (0.7 %) scat sample from porcupine, and two (1.4%) from rhesus monkey. Misidentification rate was highest for Asian palm civet (25.7%), followed by red fox (11.1%) and Asiatic jackal (9.7%) but least for common leopard scats (4.2%). The results specific to our study area concur with previous studies that have recommended that carnivore monitoring programs utilize molecular identification of predator scats. Using only morphological identification of scats can be misleading and may result in wrong management decisions.
DNA metabarcoding for diet analysis and biodiversity: A case study using the endangered Australian sea lion (Neophoca cinerea)
DOI:10.1002/ece3.3123
PMID:28770080
[本文引用: 1]

The analysis of apex predator diet has the ability to deliver valuable insights into ecosystem health, and the potential impacts a predator might have on commercially relevant species. The Australian sea lion () is an endemic apex predator and one of the world's most endangered pinnipeds. Given that prey availability is vital to the survival of top predators, this study set out to understand what dietary information DNA metabarcoding could yield from 36 sea lion scats collected across 1,500 km of its distribution in southwest Western Australia. A combination of PCR assays were designed to target a variety of potential sea lion prey, including mammals, fish, crustaceans, cephalopods, and birds. Over 1.2 million metabarcodes identified six classes from three phyla, together representing over 80 taxa. The results confirm that the Australian sea lion is a wide-ranging opportunistic predator that consumes an array of mainly demersal fauna. Further, the important commercial species (southern calamari squid) and (western rock lobster) were detected, but were present in <25% of samples. Some of the taxa identified, such as fish, sharks and rays, clarify previous knowledge of sea lion prey, and some, such as eel taxa and two gastropod species, represent new dietary insights. Even with modest sample sizes, a spatial analysis of taxa and operational taxonomic units found within the scat shows significant differences in diet between many of the sample locations and identifies the primary taxa that are driving this variance. This study provides new insights into the diet of this endangered predator and confirms the efficacy of DNA metabarcoding of scat as a noninvasive tool to more broadly define regional biodiversity.
Comparison of diet and prey selectivity of the Pyrenean desman and the Eurasian water shrew using next-generation sequencing methods
DOI:10.1016/j.mambio.2017.09.001 URL [本文引用: 1]
Who’s for dinner? High-throughput sequencing reveals bat dietary differentiation in a biodiversity hotspot where prey taxonomy is largely undescribed
DOI:10.1111/mec.12531 URL [本文引用: 1]
Distribution and food habits of mammals in the vicinity of the Reelfoot Lake Biological Station
Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms
DOI:10.1038/ismej.2012.8 [本文引用: 1]
Variations in food habit and viscera organ morphology of four rodents in Shennongjia, central China
神农架地区4种啮齿类食性及脏器形态差异比较
A note on the diet of the European Water shrew, Neomys fodiens bicolor
DOI:10.1111/jzo.1979.188.issue-2 URL [本文引用: 1]
Diets and coexistence in Neomys and Sorex shrews in Białowieża forest, eastern Poland
DOI:10.1111/jzo.2006.269.issue-3 URL [本文引用: 2]
Food niche overlap and ecological separation in a multi-species community of shrews in the Siberian taiga
DOI:10.1111/jzo.1994.234.issue-1 URL [本文引用: 1]
The diet of Myotis lucifugus across Canada: Assessing foraging quality and diet variability
Deciphering the isotopic niches of now-extinct Hispaniolan rodents
DOI:10.1080/02724634.2018.1510414 URL [本文引用: 2]
Observations on the food and feeding of the angler-fish, Lophim piscatorius L., in the northern Irish Sea
DOI:10.1111/jfb.1985.27.issue-5 URL [本文引用: 1]
Darwin’s finches and their diet niches: The sympatric coexistence of imperfect generalists
DOI:10.1111/jeb.12383
PMID:24750315
[本文引用: 1]

Adaptive radiation can be strongly influenced by interspecific competition for resources, which can lead to diverse outcomes ranging from competitive exclusion to character displacement. In each case, sympatric species are expected to evolve into distinct ecological niches, such as different food types, yet this expectation is not always met when such species are examined in nature. The most common hypotheses to account for the coexistence of species with substantial diet overlap rest on temporal variation in niches (often diets). Yet spatial variation in niche overlap might also be important, pointing to the need for spatiotemporal analyses of diet and diet overlap between closely related species persisting in sympatry. We here perform such an analysis by characterizing the diets of, and diet overlap among, four sympatric Darwin's ground finch species at three sites and over 5 years on a single Galápagos island (Santa Cruz). We find that the different species have broadly similar and overlapping diets - they are to some extent generalists and opportunists - yet we also find that each species retains some 'private' resources for which their morphologies are best suited. Importantly, use of these private resources increased considerably, and diet overlap decreased accordingly, when the availability of preferred shared foods, such as arthropods, was reduced during drought conditions. Spatial variation in food resources was also important. These results together suggest that the ground finches are 'imperfect generalists' that use overlapping resources under benign conditions (in space or time), but then retreat to resources for which they are best adapted during periods of food limitation. These conditions likely promote local and regional coexistence.© 2014 The Authors. Journal of Evolutionary Biology © 2014 European Society For Evolutionary Biology.
Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data?
DOI:10.1111/mec.14734
PMID:29858539
[本文引用: 1]

Advances in DNA sequencing technology have revolutionized the field of molecular analysis of trophic interactions, and it is now possible to recover counts of food DNA sequences from a wide range of dietary samples. But what do these counts mean? To obtain an accurate estimate of a consumer's diet should we work strictly with data sets summarizing frequency of occurrence of different food taxa, or is it possible to use relative number of sequences? Both approaches are applied to obtain semi-quantitative diet summaries, but occurrence data are often promoted as a more conservative and reliable option due to taxa-specific biases in recovery of sequences. We explore representative dietary metabarcoding data sets and point out that diet summaries based on occurrence data often overestimate the importance of food consumed in small quantities (potentially including low-level contaminants) and are sensitive to the count threshold used to define an occurrence. Our simulations indicate that using relative read abundance (RRA) information often provides a more accurate view of population-level diet even with moderate recovery biases incorporated; however, RRA summaries are sensitive to recovery biases impacting common diet taxa. Both approaches are more accurate when the mean number of food taxa in samples is small. The ideas presented here highlight the need to consider all sources of bias and to justify the methods used to interpret count data in dietary metabarcoding studies. We encourage researchers to continue addressing methodological challenges and acknowledge unanswered questions to help spur future investigations in this rapidly developing area of research.© 2018 John Wiley & Sons Ltd.
Harm of rodents to human health
啮齿动物对人类健康的危害
UPARSE: Highly accurate OTU sequences from microbial amplicon reads
DOI:10.1038/nmeth.2604
PMID:23955772
[本文引用: 1]

Amplified marker-gene sequences can be used to understand microbial community structure, but they suffer from a high level of sequencing and amplification artifacts. The UPARSE pipeline reports operational taxonomic unit (OTU) sequences with ≤1% incorrect bases in artificial microbial community tests, compared with >3% incorrect bases commonly reported by other methods. The improved accuracy results in far fewer OTUs, consistently closer to the expected number of species in a community.
Winter habitat characteristics of Sichuan partridge (Arborophila rufipectus)
四川山鹧鸪冬季栖息地特征
四川山鹧鸪(Arborophila rufipectus)是我国西南山地特有的珍稀濒危鸟类,迄今对其越冬栖息地的了解甚少。2013年1月和2014年1—2月,在四川老君山国家级自然保护区对其冬季栖息地特征进行了研究。野外共测定了48个利用样方和30个对照样方的特征参数。结果表明:四川山鹧鸪偏好选择海拔较低、坡度平缓、乔木盖度较小、竹子较矮、草本低矮稀疏、1.0 m层盖度较大、4.0~5.0 m层盖度较小、距小路和林缘较近的栖息地。此外,四川山鹧鸪还倾向于选择在距水源较近及地表落叶丰富的栖息地活动。安全与食物条件可能是影响四川山鹧鸪冬季栖息地选择的关键因素。
The niche-relationships of the California thrasher
DOI:10.2307/4072271 URL [本文引用: 1]
Applications of DNA metabarcoding in diet identification of herbivores
DNA宏条形码技术在食草动物食性研究中的应用
DOI:10.13560/j.cnki.biotech.bull.1985.2020-0988
[本文引用: 1]

随着测序技术的快速发展,整合DNA条形码和高通量测序的DNA宏条形码技术已经成为当前研究热点之一,在食草动物的食性鉴定中有很大潜力。放牧动物食性研究是动物营养学和草地生态学领域的重要研究内容。而与传统食性研究方法相比,宏条形码技术可通过对植物DNA条形码的高通量测序,获得样本中的物种组成进而分析动物食性。介绍了传统食性分析手段的局限,重点综述了DNA宏条形码技术的产生、操作原理以及在食草类动物食性鉴定领域中的应用,同时还简述了可能存在的挑战,并对该技术今后的发展方向进行了展望。
The competitive exclusion principle
DOI:10.1126/science.131.3409.1292 PMID:14399717 [本文引用: 1]
A multi-locus phylogeny of Nectogalini shrews and influences of the paleoclimate on speciation and evolution
DOI:10.1016/j.ympev.2010.03.039
PMID:20363345
[本文引用: 1]

Nectogaline shrews are a major component of the small mammalian fauna of Europe and Asia, and are notable for their diverse ecology, including utilization of aquatic habitats. So far, molecular phylogenetic analyses including nectogaline species have been unable to infer a well-resolved, well-supported phylogeny, thus limiting the power of comparative evolutionary and ecological analyses of the group. Here, we employ Bayesian phylogenetic analyses of eight mitochondrial and three nuclear genes to infer the phylogenetic relationships of nectogaline shrews. We subsequently use this phylogeny to assess the genetic diversity within the genus Episoriculus, and determine whether adaptation to aquatic habitats evolved independently multiple times. Moreover, we both analyze the fossil record and employ Bayesian relaxed clock divergence dating analyses of DNA to assess the impact of historical global climate change on the biogeography of Nectogalini. We infer strong support for the polyphyly of the genus Episoriculus. We also find strong evidence that the ability to heavily utilize aquatic habitats evolved independently in both Neomys and Chimarrogale+Nectogale lineages. Our Bayesian molecular divergence analysis suggests that the early history of Nectogalini is characterized by a rapid radiation at the Miocene/Pliocene boundary, thus potentially explaining the lack of resolution at the base of the tree. Finally, we find evidence that nectogalines once inhabited northern latitudes, but the global cooling and desiccating events at the Miocene/Pliocene and Pliocene/Pleistocene boundaries and Pleistocene glaciation resulted in the migration of most Nectogalini lineages to their present day southern distribution.Copyright 2010 Elsevier Inc. All rights reserved.
Diet and ectoparasites of the southern short-tailed shrew (Blarina carolinensis) in Louisiana
DOI:10.3398/064.072.0414 URL [本文引用: 1]
A molecular analysis of ground sloth diet through the last glaciation
DNA was extracted from five coprolites, excavated in Gypsum Cave, Nevada and radiocarbon dated to approximately 11 000, 20 000 and 28 500 years BP. All coprolites contained mitochondrial DNA sequences identical to a DNA sequence determined from a bone of the extinct ground sloth Nothrotheriops shastensis. A 157-bp fragment of the chloroplast gene for the large subunit of the ribulosebisphosphate carboxylase (rbcL) was amplified from the boluses and several hundred clones were sequenced. In addition, the same DNA fragment was sequenced from 99 plant species that occur in the vicinity of Gypsum Cave today. When these were compared to the DNA sequences in GenBank, 69 were correctly (two incorrectly) assigned to taxonomic orders. The plant sequences from the five coprolites as well as from one previously studied coprolite were compared to rbcL sequences in GenBank and the contemporary plant species. Thirteen families or orders of plants that formed part of the diet of the Shasta ground sloth could be identified, showing that the ground sloth was feeding on trees as well as herbs and grasses. The plants in the boluses further indicate that the climate 11 000 years BP was dryer than 20 000 and 28 500 years BP. However, the sloths seem to have visited water sources more frequently at 11 000 BP than at earlier times.
Influences of submerged plant collapse on diet composition, breadth, and overlap among four crane species at Poyang Lake, China
DOI:10.1186/s12983-020-00382-w
[本文引用: 2]

Condition indices (CIs) are used in ecological studies as a way of measuring an individual animal’s health and fitness. Noninvasive CIs are estimations of a relative score of fat content or rely on a ratio of body mass compared to some measure of size, usually a linear dimension such as tarsus or snout-vent length. CIs are generally validated invasively by lethal fat extraction as in a seasonal sample of individuals in a population. Many alternatives to lethal fat extraction are costly or time consuming. As an alternative, dual-energy X-ray absorptiometry (DXA) allows for non-destructive analysis of body composition and enables multiple measurements during an animal’s life time. DXA has never been used for ecological studies in a small, free-ranging lizard before, therefore we calibrated this method against a chemical extraction of fat from a sample of 6 geckos (Israeli fan toed gecko Ptyodactylus guttatus) ranging in body mass between 4.2–11.5 g. We then used this calibrated DXA measurements to determine the best linear measurement calculated CI for this species.
Biodiversity of plankton by species oscillations and chaos
Food and ectoparasites of the southern short-tailed shrew, Blarina carolinensis (Mammalia: Soricidae), from South Carolina
An improved method for utilizing high-throughput amplicon sequencing to determine the diets of insectivorous animals
DOI:10.1111/1755-0998.12951
PMID:30281913
[本文引用: 1]

DNA analysis of predator faeces using high-throughput amplicon sequencing (HTS) enhances our understanding of predator-prey interactions. However, conclusions drawn from this technique are constrained by biases that occur in multiple steps of the HTS workflow. To better characterize insectivorous animal diets, we used DNA from a diverse set of arthropods to assess PCR biases of commonly used and novel primer pairs for the mitochondrial gene, cytochrome oxidase C subunit 1 (COI). We compared diversity recovered from HTS of bat guano samples using a commonly used primer pair "ZBJ" to results using the novel primer pair "ANML." To parameterize our bioinformatics pipeline, we created an arthropod mock community consisting of single-copy (cloned) COI sequences. To examine biases associated with both PCR and HTS, mock community members were combined in equimolar amounts both pre- and post-PCR. We validated our system using guano from bats fed known diets and using composite samples of morphologically identified insects collected in pitfall traps. In PCR tests, the ANML primer pair amplified 58 of 59 arthropod taxa (98%), whereas ZBJ amplified 24-40 of 59 taxa (41%-68%). Furthermore, in an HTS comparison of field-collected samples, the ANML primers detected nearly fourfold more arthropod taxa than the ZBJ primers. The additional arthropods detected include medically and economically relevant insect groups such as mosquitoes. Results revealed biases at both the PCR and sequencing levels, demonstrating the pitfalls associated with using HTS read numbers as proxies for abundance. The use of an arthropod mock community allowed for improved bioinformatics pipeline parameterization.© 2018 John Wiley & Sons Ltd.
DNA metabarcoding illuminates dietary niche partitioning by African large herbivores
An integrative approach to detect subtle trophic niche differentiation in the sympatric trawling bat species Myotis dasycneme and Myotis daubentonii
DOI:10.1111/mec.12512
PMID:24164379
[本文引用: 1]

Bats are well known for species richness and ecological diversity, and thus, they provide a good opportunity to study relationships and interaction between species. To assess interactions, we consider distinct traits that are probably to be triggered by niche shape and evolutionary processes. We present data on the trophic niche differentiation between two sympatric European trawling bat species, Myotis dasycneme and Myotis daubentonii, incorporating a wide spectrum of methodological approaches. We measure morphological traits involved in foraging and prey handling performance including bite force, weightlifting capacity and wing morphology. We then measure resulting prey consumption using both morphological and molecular diet analyses. These species closely resemble each other in morphological traits, however, subtle but significant differences were apparent in bite force and lift capacity, which are related to differences in basic body and head size. Both morphological and molecular diet analyses show strong niche overlap. We detected subtle differences in less frequent prey items, as well as differences in the exploitation of terrestrial and aquatic-based prey groups. Myotis dasycneme feeds more on aquatic prey, like Chironomidae and their pupal stages, or on the aquatic moth Acentria ephemerella. Myotis daubentonii feeds more on terrestrial prey, like Brachycera, or Coleoptera. This suggests that these bats use different microhabitats within the habitat where they co-occur.© 2013 John Wiley & Sons Ltd.
Food habits of the southern short-tailed shrew (Blarina carolinensis) in East Texas
Analysis of feeding habits of forest rodents in Wangwu Mountain area of Jiyuan City
济源市王屋山地区林栖鼠类食性分析
Preliminary observation on feeding habits and activities of Rattus norvegicus and Rattus flavipectus
对褐家鼠、黄胸鼠食性及活动规律的初步观察
Habitat use by endangered Sichuan partridges Arborophila rufipectus during the breeding season
DOI:10.3161/000164508X395298 URL [本文引用: 1]
Habitat utilization of the Sichuan hill partridge (Arborophila rufipectus) in the non-breeding period in Laojunshan Nature Reserve
The application of high-throughput sequencing technologies to wildlife diet analysis
高通量测序技术在野生动物食性分析中的应用
Food habits of blue sheep, Pseudois nayaur in the Helan Mountains, China
Metabarcoding diet analysis of snow leopards (Panthera uncia) in Wolong National Nature Reserve, Sichuan Province
DOI:10.17520/biods.2019101
[本文引用: 3]

As the apex predator of plateau ecosystems in Central Asia and the Qinghai-Tibet Plateau, the snow leopard (Panthera uncia) plays an essential role in maintaining food-web structure and ecosystem stability. Learning the diet composition and dynamics of the snow leopard is important for understanding its role in ecosystem functioning and interspecific interactions. Previous diet analyses of the snow leopard have been based mainly on morphological identification of food debris in the feces, though the accuracy of this practice has been broadly debated. The Qionglai Mountains are located at the southeast edge of the snow leopard range, harboring a small and relatively isolated population of snow leopards that are barely studied. Using non-invasive sampling, we collected 38 putative snow leopard fecal samples in the Wolong National Nature Reserve in the Qionglai Mountains. To identify the fecal origin, we extracted the fecal DNA and amplified the mitochondrial DNA 16S rRNA gene fragment. Twenty-two fecal samples were identified as originating from snow leopards. Subsequently, vertebrate universal primers and a snow leopard-specific blocking oligo were used to amplify the food components in the fecal DNA, and then high-throughput sequencing was performed to analyze the diet composition of snow leopards. The blue sheep (Pseudois nayaur) was detected in 67% of the samples and was found to be the main staple food of snow leopards’ diet. The domestic yak (Bos grunniens) appeared in 33% of the fecal samples, also accounting for a high proportion of the snow leopard diet. In addition, pikas (Ochotona spp.) and birds were found in a small number of fecal samples. Therefore, wild prey was found to be the main food source for snow leopards in Wolong. However, livestock (yak) also accounted for a relatively large proportion of their diet.
基于分子宏条形码分析四川卧龙国家级自然保护区雪豹的食性
DOI:10.17520/biods.2019101
[本文引用: 3]

作为中亚和青藏高原山地生态系统中的顶级捕食者, 雪豹(Panthera uncia)对于维持食物网结构和生态系统稳定性有重要作用。了解雪豹的食性组成和变化对于理解其生态系统功能和物种间相互作用有重要意义。以往的雪豹食性分析多基于对其粪便中食物残渣的形态学鉴定, 但准确度受人员经验和主观因素影响较大。邛崃山脉位于雪豹分布区东南缘, 该区域的雪豹种群规模小且相对孤立, 研究匮乏。本研究基于非损伤性取样, 在邛崃山脉的卧龙国家级保护区采集疑似雪豹粪便样品38份, 首先提取粪便DNA, 并扩增线粒体DNA 16S rRNA基因片段进行分子物种鉴定, 确定其中22份为雪豹粪便样品。随后, 利用脊椎动物通用引物和雪豹特异性阻抑引物扩增粪便DNA中的食物成分, 并进行高通量测序, 分析雪豹食性构成。食性分析结果显示岩羊(Pseudois nayaur)是卧龙地区雪豹最主要的食物, 在67%的样品中均有检出。家牦牛(Bos grunniens)在33%的样品中出现, 也在雪豹食性中占较高比例。此外, 鼠兔(Ochotona spp.)和鸟类也在少量样品中发现。可见, 野生猎物是卧龙地区雪豹的主要食物资源; 与大世界大多数其他地区的雪豹食性相同, 野生大型有蹄类是雪豹最重要的食物。然而家畜(牦牛)在卧龙雪豹食谱中有相当高的占比, 显示该区域内可能存在较为严重的由雪豹捕食散养家畜引起的人兽冲突问题。
On optimal use of a patchy environment
DOI:10.1086/282454 URL [本文引用: 1]
Evidence for interspecific competition between feral ass Equus asinus and mountain sheep Ovis canadensis in a desert environment
DOI:10.2981/0909-6396(2008)14[228:EFICBF]2.0.CO;2 URL [本文引用: 1]
The structure of lizard communities
DOI:10.1146/ecolsys.1973.4.issue-1 URL [本文引用: 1]
Who is eating what: Diet assessment using next generation sequencing
DOI:10.1111/j.1365-294X.2011.05403.x
PMID:22171763
[本文引用: 2]

The analysis of food webs and their dynamics facilitates understanding of the mechanistic processes behind community ecology and ecosystem functions. Having accurate techniques for determining dietary ranges and components is critical for this endeavour. While visual analyses and early molecular approaches are highly labour intensive and often lack resolution, recent DNA-based approaches potentially provide more accurate methods for dietary studies. A suite of approaches have been used based on the identification of consumed species by characterization of DNA present in gut or faecal samples. In one approach, a standardized DNA region (DNA barcode) is PCR amplified, amplicons are sequenced and then compared to a reference database for identification. Initially, this involved sequencing clones from PCR products, and studies were limited in scale because of the costs and effort required. The recent development of next generation sequencing (NGS) has made this approach much more powerful, by allowing the direct characterization of dozens of samples with several thousand sequences per PCR product, and has the potential to reveal many consumed species simultaneously (DNA metabarcoding). Continual improvement of NGS technologies, on-going decreases in costs and current massive expansion of reference databases make this approach promising. Here we review the power and pitfalls of NGS diet methods. We present the critical factors to take into account when choosing or designing a suitable barcode. Then, we consider both technical and analytical aspects of NGS diet studies. Finally, we discuss the validation of data accuracy including the viability of producing quantitative data.© 2011 Blackwell Publishing Ltd.
Dietary differences among sympatric Callitrichinae in northern Bolivia: Callimico goeldii, Saguinus fuscicollis and S. labiatus
DOI:10.1023/A:1012013621258 URL [本文引用: 1]
High-throughput sequencing offers insight into mechanisms of resource partitioning in cryptic bat species
DOI:10.1002/ece3.49
PMID:22393522
[本文引用: 1]

Sympatric cryptic species, characterized by low morphological differentiation, pose a challenge to understanding the role of interspecific competition in structuring ecological communities. We used traditional (morphological) and novel molecular methods of diet analysis to study the diet of two cryptic bat species that are sympatric in southern England (Plecotus austriacus and P. auritus) (Fig. 1). Using Roche FLX 454 (Roche, Basel, CH) high-throughput sequencing (HTS) and uniquely tagged generic arthropod primers, we identified 142 prey Molecular Operational Taxonomic Units (MOTUs) in the diet of the cryptic bats, 60% of which were assigned to a likely species or genus. The findings from the molecular study supported the results of microscopic analyses in showing that the diets of both species were dominated by lepidopterans. However, HTS provided a sufficiently high resolution of prey identification to determine fine-scale differences in resource use. Although both bat species appeared to have a generalist diet, eared-moths from the family Noctuidae were the main prey consumed. Interspecific niche overlap was greater than expected by chance (O(jk) = 0.72, P < 0.001) due to overlap in the consumption of the more common prey species. Yet, habitat associations of nongeneralist prey species found in the diets corresponded to those of their respective bat predator (grasslands for P. austriacus, and woodland for P. auritus). Overlap in common dietary resource use combined with differential specialist prey habitat associations suggests that habitat partitioning is the primary mechanism of coexistence. The performance of HTS is discussed in relation to previous methods of molecular and morphological diet analysis. By enabling species-level identification of dietary components, the application of DNA sequencing to diet analysis allows a more comprehensive comparison of the diet of sympatric cryptic species, and therefore can be an important tool for determining fine-scale mechanisms of coexistence.
Vsearch: A versatile open source tool for metagenomics
DOI:10.7717/peerj.2584
URL
[本文引用: 1]

VSEARCH is an open source and free of charge multithreaded 64-bit tool for processing and preparing metagenomics, genomics and population genomics nucleotide sequence data. It is designed as an alternative to the widely used USEARCH tool (Edgar, 2010) for which the source code is not publicly available, algorithm details are only rudimentarily described, and only a memory-confined 32-bit version is freely available for academic use.
Reconstruction of the historical range alters niche estimates in an endangered rodent
DOI:10.1111/ecog.04238
[本文引用: 1]

Defining historical baselines is critical for species conservation. Under the niche reduction hypothesis, species in decline may be restricted disproportionately from parts of their environmental niche. This bias likely has important implications for modeling species' distributions if only contemporary occurrences (i.e. post-range reduction) are used, because suitable habitat will be classified as unsuitable. Unfortunately, robust historical occurrence data is rarely available for sensitive species. In this study, we documented historical locations of the endangered, keystone giant kangaroo rat Dipodomys ingens by examining aerial imagery for burrow mounds. These burrow mounds are readily identifiable and distinguishable from other soil disturbances. We found giant kangaroo rat burrows well outside the currently accepted estimate of their historical distribution. Following the niche reduction hypothesis, we found that giant kangaroo rats have been extirpated from the flattest, hottest, driest parts of their range due to agricultural conversion. This reduction in their realized niche led to significant changes between historical and contemporary models of their distribution. We found that giant kangaroo rats may have occupied up to 56% more habitat historically than currently believed. Our results provide new guidance for managers working on restoration and habitat protection for this ecosystem engineer. This study highlights the critical importance of modeling historical distributions using the entire environmental niche once occupied by species of conservation need.
Prey partitioning and livestock consumption in the world’s richest large carnivore assemblage
DOI:10.1016/j.cub.2021.08.067 URL [本文引用: 1]
Fast surveys and molecular diet analysis of carnivores based on fecal DNA and metabarcoding
DOI:10.17520/biods.2018214
[本文引用: 1]

Large carnivores play an important role in the regulation of food-web structure and ecosystem functioning. However, large carnivores face serious threats that have caused declines in their populations and geographic ranges due to habitat loss and degradation, hunting, human disturbance and pathogen transmission. Conservation of large carnivore species richness and population size has become a pressing issue and an important research focus of conservation biology. The western Sichuan Plateau, located at the intersection of the mountains of southwest China and the eastern margin of the Tibetan Plateau, is a global biodiversity hotspot and has high carnivore species richness. However, increasing human activities may exacerbate the destruction of local flora and fauna, thereby threatening the survival of wild carnivores. Information on species composition and dietary habits can improve our understanding of the structure and function of the ecosystem and food-web relationships in the study area. In addition, species composition and dietary habits are of great significance for understanding multi-species coexistence mechanisms and preserving biodiversity. This study collected carnivore fecal samples from Xinlong and Shiqu counties in the Ganzi Tibetan Autonomous Prefecture, Sichuan Province. DNA was then extracted from the samples and the species was identified based on DNA sequences and DNA barcoding techniques. Seven carnivores were identified, including five large carnivores (Canis lupus, Ursus arctos, Panthera pardus, P. uncia and Canis lupus familiaris) and two medium and small-sized carnivores (Prionailurus bengalensis and Vulpes vulpes). Using fecal DNA, high-throughput sequencing and metabarcoding, we conducted diet analysis for the seven carnivores and found 28 different food molecular operational taxonomic units (MOTUs), including 19 mammals, eight birds and one fish species. The predominant prey categories of wolves, dogs and brown bears were ungulates. The domestic yak (Bos grunniens) was the most frequently identified prey species. Small mammals such as rodents and lagomorphs accounted for a significant proportion in the diets of leopard cats and red foxes, The most frequent prey of this category of carnivore were the Chinese scrub vole (Neodon irene) and plateau pika (Ochotona curzoniae). In addition, leopards and snow leopards mainly fed on the Chinese goral (Naemorhedus griseus) and blue sheep (Pseudois nayaur), respectively. Our study highlights the utility of fecal DNA and metabarcoding technique in fast carnivore surveys and high-throughput diet analysis, and provides a technical reference and guidance for future biodiversity surveys and food-web studies.
基于粪便DNA及宏条形码技术的食肉动物快速调查及食性分析
DOI:10.17520/biods.2018214
[本文引用: 1]

在陆地生态系统中, 大型食肉动物对于稳定食物网结构和生态系统功能有重要作用。在世界范围内, 由于栖息地丧失和破碎化、猎杀、人类活动干扰以及病原体的传播, 大型食肉动物生存正面临严重威胁, 多种食肉动物地理分布范围及种群数量大幅度缩减。如何有效保护大型食肉动物物种多样性及种群已经成为世界关注的焦点问题和保护生物学的重要研究方向。川西高原地处我国西南山地与青藏高原东缘交界地带, 属于世界生物多样性热点地区, 是世界大型食肉动物物种最丰富的地区之一, 而日益增强的人类活动可能会加剧对当地动植物资源的破坏, 进而威胁野生食肉动物的生存。获得准确的物种多样性信息及食肉动物食性数据有助于深入了解该地区生态系统结构及食物网关系, 对研究物种共存机制及生物多样性保护有重要意义。本研究通过从四川甘孜藏族自治州新龙县和石渠县野外采集的食肉动物粪便样品中提取DNA, 利用DNA条形码进行物种鉴定, 快速获得该地区食肉动物物种构成信息。38份粪便样品经鉴定来自于7种食肉动物, 分别为5种大型食肉动物(狼Canis lupus、棕熊Ursus arctos、豹Panthera pardus、雪豹P. unica、狗Canis lupus familiaris)和2种中小型食肉动物(豹猫Prionailurus bengalensis、赤狐Vulpes vulpes)。进一步利用高通量测序和宏条形码技术对7种食肉动物粪便中的食物DNA进行精准食性分析, 得到包含19种哺乳类、8种鸟类和1种鱼类共计28个不同的食物分子可操作分类单元(molecular operational taxonomic unit, MOTU)。结果显示, 狼、狗、棕熊最主要的食物来源为偶蹄目动物, 其中取食频率最高的物种为家牦牛(Bos grunniens); 而豹猫和赤狐食物中小型哺乳动物如啮齿目和兔形目占重要比例, 其中高原松田鼠(Neodon irene)和高原鼠兔(Ochotona curzoniae)被取食频率最高。豹和雪豹的食物分别为偶蹄目的中华斑羚(Naemorhedus griseus)和岩羊(Pseudois nayaur)。本研究显示了粪便DNA及宏条形码技术在食肉动物多样性快速调查及高通量精确食性分析中的应用前景, 并为此类研究提供了技术路线的有力借鉴。
Gradients in richness and turnover of a forest passerine’s diet prior to breeding: A mixed model approach applied to faecal metabarcoding data
DOI:10.1111/mec.15394
PMID:32100904
[本文引用: 1]

Rather little is known about the dietary richness and variation of generalist insectivorous species, including birds, due primarily to difficulties in prey identification. Using faecal metabarcoding, we provide the most comprehensive analysis of a passerine's diet to date, identifying the relative magnitudes of biogeographic, habitat and temporal trends in the richness and turnover in diet of Cyanistes caeruleus (blue tit) along a 39 site and 2° latitudinal transect in Scotland. Faecal samples were collected in 2014-2015 from adult birds roosting in nestboxes prior to nest building. DNA was extracted from 793 samples and we amplified COI and 16S minibarcodes. We identified 432 molecular operational taxonomic units that correspond to putative dietary items. Most dietary items were rare, with Lepidoptera being the most abundant and taxon-rich prey order. Here, we present a statistical approach for estimation of gradients and intersample variation in taxonomic richness and turnover using a generalised linear mixed model. We discuss the merits of this approach over existing tools and present methods for model-based estimation of repeatability, taxon richness and Jaccard indices. We found that dietary richness increases significantly as spring advances, but changes little with elevation, latitude or local tree composition. In comparison, dietary composition exhibits significant turnover along temporal and spatial gradients and among sites. Our study shows the promise of faecal metabarcoding for inferring the macroecology of food webs, but we also highlight the challenge posed by contamination and make recommendations of laboratory and statistical practices to minimise its impact on inference.© 2020 John Wiley & Sons Ltd.
Identifying the diet of a declining prairie grouse using DNA metabarcoding
DOI:10.1642/AUK-17-199.1 URL [本文引用: 1]
Towards next-generation biodiversity assessment using DNA metabarcoding
DOI:10.1111/j.1365-294X.2012.05470.x
PMID:22486824
[本文引用: 1]

Virtually all empirical ecological studies require species identification during data collection. DNA metabarcoding refers to the automated identification of multiple species from a single bulk sample containing entire organisms or from a single environmental sample containing degraded DNA (soil, water, faeces, etc.). It can be implemented for both modern and ancient environmental samples. The availability of next-generation sequencing platforms and the ecologists' need for high-throughput taxon identification have facilitated the emergence of DNA metabarcoding. The potential power of DNA metabarcoding as it is implemented today is limited mainly by its dependency on PCR and by the considerable investment needed to build comprehensive taxonomic reference libraries. Further developments associated with the impressive progress in DNA sequencing will eliminate the currently required DNA amplification step, and comprehensive taxonomic reference libraries composed of whole organellar genomes and repetitive ribosomal nuclear DNA can be built based on the well-curated DNA extract collections maintained by standardized barcoding initiatives. The near-term future of DNA metabarcoding has an enormous potential to boost data acquisition in biodiversity research.© 2012 Blackwell Publishing Ltd.
DNA metabarcoding provides insights into seasonal diet variations in Chinese mole shrew (Anourosorex squamipes) with potential implications for evaluating crop impacts
DOI:10.1002/ece3.v11.1 URL [本文引用: 3]
Notes on the ecological concepts of habitat, biotope and niche
DOI:10.2307/1929830 URL [本文引用: 1]
Spatial distribution of sympatric Rhinopithecus brelichi and Macaca thibetana in Fanjingshan National Nature Reserve, Guizhou, China
梵净山保护区同域分布黔金丝猴与藏酋猴的空间分布初探
The ecological significance and controlling of rodent outbreaks in the Qinghai-Tibetan Grasslands
青藏高原草地鼠害的生态释义及控制
Causes and control countermeasures of forest rodent damage in Bailongjiang forest area
白龙江林区森林鼠害发生原因及防治对策
Trophic niches of four species of common small mammals in Inner Mongolia grassland and their relationships
内蒙古典型草原4种常见小哺乳动物的营养生态位及相互关系
A study on Gansu Zokor’s feeding rhythm and its food preference to different beits
甘肃鼢鼠取食节律及对不同饵料喜食性的研究
Progress on high-throughput sequencing and its applications in food web analysis
高通量测序及其在食物网解析中的应用进展
Food of the southern short-tailed shrew (Blarina carolinensis) on Cumberland Island, Georgia
DOI:10.1656/1528-7092(2006)5[361:FOTSSS]2.0.CO;2 URL [本文引用: 1]
Habitat use and change of Arborophila rufiperctus during the breeding season in the Laojunshan National Nature Reserve, Sichuan
四川老君山国家级自然保护区四川山鹧鸪繁殖期的生境利用及其变化研究
Research on autumn time niche of five rodents in forests ecosystem
森林生态系统中5种啮齿动物秋季时间生态位
Selection of a marker gene to construct a reference library for wetland plants, and the application of metabarcoding to analyze the diet of wintering herbivorous waterbirds
DOI:10.7717/peerj.2345
URL
[本文引用: 1]

Food availability and diet selection are important factors influencing the abundance and distribution of wild waterbirds. In order to better understand changes in waterbird population, it is essential to figure out what they feed on. However, analyzing their diet could be difficult and inefficient using traditional methods such as microhistologic observation. Here, we addressed this gap of knowledge by investigating the diet of greater white-fronted gooseAnser albifronsand bean gooseAnser fabalis, which are obligate herbivores wintering in China, mostly in the Middle and Lower Yangtze River floodplain. First, we selected a suitable and high-resolution marker gene for wetland plants that these geese would consume during the wintering period. Eight candidate genes were included:rbcL,rpoC1,rpoB,matK,trnH-psbA,trnL (UAA),atpF-atpH, andpsbK-psbI. The selection was performed via analysis of representative sequences from NCBI and comparison of amplification efficiency and resolution power of plant samples collected from the wintering area. ThetrnL gene was chosen at last with c/h primers, and a local plant reference library was constructed with this gene. Then, utilizing DNA metabarcoding, we discovered 15 food items in total from the feces of these birds. Of the 15 unique dietary sequences, 10 could be identified at specie level. As for greater white-fronted goose, 73% of sequences belonged toPoaceaespp., and 26% belonged toCarexspp. In contrast, almost all sequences of bean goose belonged toCarexspp. (99%). Using the same samples, microhistology provided consistent food composition with metabarcoding results for greater white-fronted goose, while 13% ofPoaceaewas recovered for bean goose. In addition, two other taxa were discovered only through microhistologic analysis. Although most of the identified taxa matched relatively well between the two methods, DNA metabarcoding gave taxonomically more detailed information. Discrepancies were likely due to biased PCR amplification in metabarcoding, low discriminating power of current marker genes for monocots, and biases in microhistologic analysis. The diet differences between two geese species might indicate deeper ecological significance beyond the scope of this study. We concluded that DNA metabarcoding provides new perspectives for studies of herbivorous waterbird diets and inter-specific interactions, as well as new possibilities to investigate interactions between herbivores and plants. In addition, microhistologic analysis should be used together with metabarcoding methods to integrate this information.
Trophic niches of Pantholops hodgsoni, Procapra picticaudata and Equus kiang in Kekexili Region
可可西里地区藏羚羊、藏原羚和藏野驴的营养生态位
Significance and methodology of rodent’s food habit research: A review
啮齿动物食性研究的意义及方法评述
Diet composition and trophic niche characteristics of three rodents in Gannan meadow
甘南草原3种啮齿动物的食性及其营养生态位特征
DOI:10.11733/j.issn.1007-0435.2021.07.014
[本文引用: 1]

研究啮齿动物的食性及其营养生态位特征不仅可以了解高寒草甸环境变化下物种的食性适应特征,还可探讨多种啮齿动物的种间关系。本研究采用粪便显微组织观察法,分析了甘南草原高原鼠兔(Ochotona curzoniae)、高原鼢鼠(Eospalax baileyi)和喜马拉雅旱獭(Marmota himalayana)3种啮齿动物的食性及营养生态位特征。结果发现:3种啮齿动物所采食的植物种类基本相同,但其比例各异;高原鼢鼠所采食的植物主要是珠芽蓼(Polygonum viviparum)和鹅绒委陵菜(Potentilla anserina)等杂类草,且对莎草科(Cyperaceae)植物的采食量显著高于其它两种动物,高原鼠兔主要采食禾本科(Poaceae)植物,喜马拉雅旱獭则主要采食禾本科和菊科(Asteraceae)植物;营养生态位宽度依次为喜马拉雅旱獭>高原鼢鼠>高原鼠兔,其中高原鼠兔与喜马拉雅旱獭的营养生态位重叠最大(0.52)。3种啮齿动物在食物上存在分化,营养生态位分离,是其共存的重要原因。
Bibliometric analysis of food habit research based on DNA molecular biology
基于DNA分子生物学食性研究领域的文献计量分析
A review of rodent feeding methods
啮齿动物食性研究方法
Feeding and Nutrition Strategy and Feeding Habitat Evaluation of Red Deer and Sika Deer Distributed in the Same Domain
同域分布马鹿与梅花鹿采食和营养策略及采食生境评价