基因组加倍(whole-genome duplication, WGD)形成多倍体是植物进化的主要机制之一(Soltis et al, 2009; Eric Schranz et al, 2012)。几乎所有植物都经历了至少一次基因组加倍(Jiao et al, 2011), 大约35%的维管植物是多倍体(Wood et al, 2009), 很多基因组研究都发现了古老的基因组加倍事件(The Brassica rapa Genome Sequencing Project Consor- tium, 2011; Ren et al, 2018)。多倍化还是物种形成的重要驱动力, 例如十字花科植物中有大约一半是新多倍体(neopolyploids) (Hohmann et al, 2015)。然而, Mayrose等(2011)发现多倍体具有比二倍体更高的灭绝率和更低的分化速率, 大多数新形成的多倍体不能存活下来(Arrigo & Barker 2012; Soltis et al, 2014), 因此认为多倍体是植物进化的“死胡同”。持这种观点的理由包括: 多倍体基因组不稳定、有丝分裂和减数分裂不正常、少数细胞型排斥(minority cytotype exclusion, MCE)以及重复基因的有害效应等(Levin, 1975; Morgan et al, 2020), 这些因素不利于多倍体的生存。另一种观点则认为, 虽然早期的多倍体比二倍体有更高的灭绝速率, 但是建立了稳定种群的多倍体则有利于植物的进化(Soltis et al, 2014)。
多倍化事件的发生原因可能是随机的, 也有可能是受到特定环境变化事件或逆境等诱导发生的。例如, 研究发现基因组加倍和古多倍体的形成时间与一些历史灭绝事件(如白垩纪-古近纪)高度吻合, 推测古多倍体可能是由这些事件导致的(Van de Peer et al, 2017; Ren et al, 2018)。其原因有三: 一是历史灭绝事件使得多倍体占据了二倍体的生态位(Van de Peer et al, 2017); 二是古多倍体是植物在长期适应过程中无性繁殖的产物, 地下或水下的无性繁殖体躲过了流星撞击地球导致的极端气候条件(Freeling, 2017); 三是与减数分裂有关的基因大量突变富集产生未分裂的配子从而形成多倍体(Zhang et al, 2013; Freeling, 2017)。此外, 一些生物和非生物胁迫引起的自然选择也是导致多倍体形成的原因之一(Doyle & Coate, 2019; Van de Peer et al, 2021)。实际上, 有很多物种如猕猴桃、烟草、小麦、蕨类等存在多倍性自然种群(曾华等, 2009; Soltis et al, 2014; 梁思琪等, 2019), 而且大约有16%的植物物种具有混合倍性(同一物种不同倍性) (Rice et al, 2015)。例如, 在湖南雪峰山和贵州发现的野生中华猕猴桃(Actinidia chinensis)存在二倍体和四倍体, 美味猕猴桃(A. chinensis var. deliciosa)存在四倍体、五倍体和六倍体, 而且高倍性多分布于高海拔地区(曾华等, 2009; Li et al, 2010)。中华猕猴桃不同倍性杂交后发生倍性分离, 用六倍体母本与二倍体父本杂交得到的子代群体中存在从3倍体到8倍体的多倍体(饶静云等, 2012)。
多倍体进化的机制是否为植物进化的驱动力? 即多倍体及其引起的物种形成是随机发生的巧合还是某些特定环境变化或灭绝事件的产物(Van de Peer et al, 2017; Wu et al, 2020)? 研究混合倍性物种的形成与维持机制为回答这些科学问题提供了重要思路(Kolář et al, 2017)。1938年, Babcock和Stebbins提出“多倍体复合体” (polyploid complex)的概念(Babcock & Stebbins, 1938)。Stebbins (1950)在Variation and Evolution in Plants一书中对多倍性复合体进行了详细讨论, 并在Chromosomal Evolution in Higher Plants (Stebbins, 1971)中提出多倍体复合体可以作为进化单元的观点, 认为其可以为多倍体的进化研究提供准确可信的科学依据。
1 混合倍性种群的形成
自然界为什么存在混合倍性种群? 其中一个重要原因是新多倍体形成的速率高于灭绝速率(Kolář et al, 2017)。被子植物中有15%的物种形成事件起源于多倍体, 而蕨类植物中这一比例达到31% (Wood et al, 2009)。新多倍体的形成机制主要包括未减数分裂的配子、体细胞加倍和杂交等(图1) (Zhang et al, 2019)。由于种群中新产生的多倍体频率较低, 在随机交配体系中处于劣势, 无法找到适配对象, 使其被排除在外而不能固定下来, 这就是少数细胞型排斥假说(Levin, 1975)。该假说的另一种解释是不同倍性之间产生的新倍性后代(如三倍体)的适合度较低导致其不容易稳定建立种群(Kolář et al, 2017)。混合倍性种群的出现说明这些新倍性个体克服了这种少数细胞型排斥劣势, 而且确实有研究发现稀有倍性个体并没有表现出适合度劣势(Kao, 2007)。
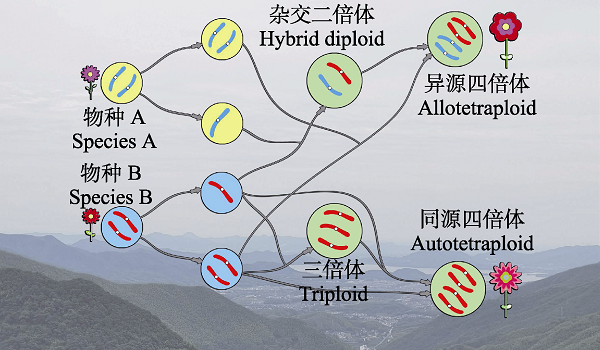
图1
图1
多倍体形成途径的简单示意图。二倍体物种产生正常的单倍体配子和未减数分裂的配子, 未减数分裂配子可以和单体配子产生三倍体, 也可以融合产生同源四倍体; 同时也可以通过体细胞加倍产生同源四倍体; 异源四倍体通过物种之间未减数分裂配子的融合或者通过物种间的杂交再加倍产生。
Fig. 1
Formation paths to polyploid species. Diploid species produce normal haploid gametes and unreduced gametes. Unreduced gametes combine with haploid gametes to give rise to triploids or fuse to be autotetraploid species. Diploids can yield somatically polyploids. Allotetraploids are yielded through the fusion of unreduced gametes or the hybridization of haploid gametes from different species.
然而, 目前仍不清楚这一少数细胞型排斥假说在形成自然混合倍性种群中有多大的作用。要克服少数细胞型排斥来形成新的多倍体种群主要有3种途径: 一是不断形成新的多倍体并随机分布来提高新多倍体的频率; 二是提高同一倍性内的交配频率而降低倍性间的交配频率; 三是提高多倍体的竞争能力和繁殖能力从而弥补依赖频率的少数细胞型排斥劣势。从目前研究结果来看, 繁殖力和可育性不能克服少数细胞型排斥, 而且在稳定环境中多倍体的竞争能力也并不高于低倍体(Kolář et al, 2017)。但是有些研究结果发现多倍体在微环境、表型、授粉媒介和病菌侵染等的生态位分化有利于其克服少数细胞型排斥的影响(Ramsey & Ramsey, 2014; Kolář et al, 2017)。此外, 由于种群间的不断迁入迁出以及人类干扰造成了空间异质性的微环境, 有利于混合倍性种群的形成(Mráz et al, 2012; Kolář et al, 2017)。例如, 混合倍性植物有65.0%的物种包含有奇数倍性的细胞型个体, 这些混合倍性种群中大约有11.6%的个体是奇数倍性的细胞型(Kolář et al, 2017)。除了特定事件导致未减数分裂的配子和体细胞加倍, 奇数倍性也可能是通过杂交形成的(Sabara et al, 2013)。稀有倍性个体的出现说明当地种群能频繁形成多倍体(Mandák et al, 2016)。这些奇数倍性和稀有倍性的出现说明自然种群中多倍体的形成经常以较高的速率出现(Ramsey & Schemske, 1998)。
2 混合倍性种群的建立与维持
多倍体形成之后能否稳定建群? 首先, 取决于其能否适应环境并成功生存下来。这个过程涉及重复基因的进化, 而基因功能的分化是物种辐射的重要机制。多倍体更高的遗传变异和遗传多样性能有效缓冲重复基因的有害效应, 通过亚功能化(sub-functionalization)和新功能化(neo-functionali- zation)分化出更多的功能特性(De Smet et al, 2013; Zhang et al, 2019), 使得多倍体比二倍体在环境适应性方面更具优势, 能更适应极端环境(Fawcett et al, 2009; Rice et al, 2019; Van de Peer et al, 2021)。有些重复基因通过甲基化沉默其表达使其避免冗余基因造成的有害效应(De Smet et al, 2013)。多倍体也会增加物种的表型多样性(Nuismer & Cunningham, 2005)。然而, 有研究发现多倍体比二倍体形成新种更慢且灭绝更快, 使其多样化速率(即物种形成和灭绝的平衡)更低(Mayrose et al, 2011)。这可能是由于多倍体的多样化速率有滞后现象。实际上, 基因组加倍常常导致更高的多样化速率(Suda et al, 2007; Van de Peer et al, 2017)。现存的有花植物大约25%-30%是新多倍体, 即目前是多倍体但还没有经过重二倍化(re-diploidization, 指多倍体亲本染色体经过重组与重排再形成二倍体的过程)形成新的物种(Suda et al, 2007; Van de Peer et al, 2017)。
其次, 取决于其与二倍体亲本种之间的生态位分化。多倍体形成之后的生态位与二倍体亲本相比, 有的更窄, 有的更宽, 更多的是占据二倍体亲本的生态位并与之重叠(Blaine Marchant et al, 2016)。这可能是由于多倍体植物比二倍体近缘种有更快的生态位分化速率(Baniaga et al, 2020)。多倍体和二倍体之间的生态位分化表现为生态位扩展或收缩, 占据父母本的中间生态位以及开拓新的生态位等(Blaine Marchant et al, 2016; Castro et al, 2020)。而且在胁迫环境中, 多倍体比二倍体更有优势, 更能抵抗干旱、低温(Folk et al, 2020; Gunn et al, 2020; Wu et al, 2020)和病虫害(Hias et al, 2018; Wang et al, 2018)等胁迫。多倍体在胁迫环境中表现的这些抗性优势使其在与二倍体亲本的竞争中占据优势(Baduel et al, 2018; Van de Peer et al, 2021), 并在胁迫环境中表现出更高的适合度和生存优势(Stevens et al, 2020)。而且, 混合倍性种群能调节自然选择压力, 增加分化选择压力, 提高不同倍性个体之间的生态位分化。同域分布的多倍体之间的分化强于异域分布的多倍体(Nuismer & Cunningham, 2005; Sonnleitner et al, 2016), 但是同域分布和异域分布的环境有明显差异。
3 混合倍性物种
为什么要选择混合倍性物种研究多倍体进化机制? 因为在混合倍性物种中能区分基因组加倍对物种表型、生态适应和基因组分化等方面的直接影响和间接影响。一般情况下, 现存的二倍体和多倍体之间的差异被认为是基因组加倍的直接影响, 但是基因组加倍后的选择压力也是二倍体和多倍体之间差异的重要原因。对近期自然形成的多倍体进行表型和基因组分析能评估基因组加倍的直接影响, 但来源于杂交的异源多倍体不能区分基因组加倍的最初影响, 而同源多倍体则没有基因组加倍的最初影响。而且混合倍性物种作为进化的中间环节, 可以反映多倍体的进化过程(Stebbins, 1971)。因为植物由低倍性到高倍性的进化趋势是基本共识, 因此可以利用多倍体复合体研究多倍体进化历史, 也可以通过杂交、物理或化学等实验手段进行人工诱导形成多倍体, 重演多倍体物种的形成过程。
基因组加倍后, 自然选择压力怎样影响多倍体植物的繁殖和适合度? 虽然多倍体植物借助更快的生态位分化来适应自然选择压力(Baniaga et al, 2020), 但是自然选择压力仍然可以通过影响基因连锁、亚基因组的重组以及基因功能分化、转座子(transposon elements)甲基化等影响多倍体植物的生存(De Smet et al, 2013; Bottani et al, 2018; Zhang et al, 2019; Wu et al, 2020)。这种影响受增加的染色体数量和基因数量影响, 而且同源多倍体和异源多倍体的影响机制不同。同源多倍体的基因连锁分析和异源多倍体的亚基因组重组分析能解析出自然选择影响多倍体植物的信号, 分析自然选择的影响途径(Yant et al, 2013; Zhang et al, 2013)。同源多倍体的染色体来自同一亲本基因组, 而异源多倍体来自不同的亲本基因组。所以分析在原生区形成的同源多倍体有利于明确基因组加倍的直接影响, 因为同源多倍体及其后代经历了相同的系统发育历史。
混合倍性物种有利于研究多倍体进化的机制和各个阶段, 包括多倍体的起源和形成、共存与分化等过程。因为混合倍性物种拥有独特的基因组和表型变化特征, 这些特征影响着物种的生态适应能力和种群之间的遗传分化(Kolář et al, 2017), 并可能导致新物种的形成, 特别是同域物种的形成(Wood et al, 2009), 因为不同倍性之间容易导致生殖隔离(Rieseberg & Willis, 2007)。然而, 多倍体之间的生殖隔离程度弱于二倍体与多倍体之间的生殖隔离程度(Hülber et al, 2015), 这可能是由于多倍体形成合子之后的生殖隔离程度弱于二倍体(Sutherland & Galloway, 2017)。这种不完全的生殖隔离导致不同倍性之间发生基因交流, 减缓了多倍体的物种分化, 从而形成混合倍性种群。
4 展望
为揭示混合倍性物种的形成、建立与维持机制, 可研究野生混合倍性种群的组成及其多样性、未减数分裂配子和无性繁殖等对混合倍性种群形成的贡献、重复基因的功能分化、多倍体的生态适应能力及其生态位分化机理。明确混合倍性野生种群的起源、生态适应与维持机理, 有助于探讨多倍体植物的进化机制。
未来建议从以下几方面开展研究: (1)明确种群内部倍性多样性的分布格局, 对比分析不同地区野生种群的奇数倍性、优势倍性和稀有倍性的比例, 分析导致这种格局的成因, 验证少数细胞型排斥假说在混合倍性种群形成中的作用。(2)检测不同倍性群体的配子和无性繁殖个体, 明确未减数分裂的配子和无性繁殖对混合倍性形成的贡献; 区分新形成的多倍体和已经建立的多倍体, 以及是从二倍体不断产生新的多倍体, 还是多倍体形成后分化产生新的多倍体, 明确种群内的混合倍性形成机理。(3)野生混合倍性种群的维持机制研究, 从重复基因功能分化、性别变异、生态位分化分析混合倍性种群的维持机制, 以及分析温度、降水和土壤条件对混合倍性种群的影响, 分析气候和土壤等非生物因子的生态位分化对混合倍性种群维持的贡献。
参考文献
Rarely successful polyploids and their legacy in plant genomes
DOI:10.1016/j.pbi.2012.03.010
PMID:22480430
[本文引用: 1]

Polyploidy, or whole genome duplication, is recognized as an important feature of eukaryotic genome evolution. Among eukaryotes, polyploidy has probably had the largest evolutionary impact on vascular plants where many contemporary species are of recent polyploid origin. Genomic analyses have uncovered evidence of at least one round of polyploidy in the ancestry of most plants, fueling speculation that genome duplications lead to increases in net diversity. In spite of the frequency of ancient polyploidy, recent analyses have found that recently formed polyploid species have higher extinction rates than their diploid relatives. These results suggest that despite leaving a substantial legacy in plant genomes, only rare polyploids survive over the long term and most are evolutionary dead-ends.Copyright © 2012 Elsevier Ltd. All rights reserved.
The American Species of Crepis: Their Interrelationships and Distribution as Affected by Polyploidy and Apomixis
The “Polyploid Hop”: Shifting challenges and opportunities over the evolutionary lifespan of genome duplications
DOI:10.3389/fevo.2018.00117 URL [本文引用: 1]
Polyploid plants have faster rates of multivariate niche differentiation than their diploid relatives
DOI:10.1111/ele.v23.1 URL [本文引用: 2]
Patterns of abiotic niche shifts in allopolyploids relative to their progenitors
DOI:10.1111/nph.14069
PMID:27399976
[本文引用: 2]

Polyploidy has extensive genetic, physiological, morphological, and ecological ramifications. While the patterns underlying the genetic and morphological consequences of polyploidy are being rapidly elucidated, the effects on ecological niche are still largely unknown. This study investigated 13 allopolyploid systems in North America (10 ferns and three angiosperms) using digitized natural history museum specimens. The abiotic niches of the allopolyploids were compared with those of their diploid progenitors using ecological niche modeling, niche analyses, and multivariate analyses. We identified four patterns of niche shifts through our analyses: niche expansion, niche contraction, niche intermediacy, and niche novelty. The classification of these shifts depended on the amount of niche overlap and breadth between the polyploid and progenitors. The most common niche shift was niche intermediacy in which the polyploid inhabited a geographic range between that of the progenitors and had a high degree of niche overlap. Each polyploid had at least partial geographic sympatry and abiotic niche overlap with one of its progenitors, suggesting that biotic and/or microclimate factors may play a larger role in polyploid establishment than previously hypothesized. This study provides a baseline for future comparisons of the diverse outcomes of genome merger and duplication on abiotic niche preference.© 2016 The Authors. New Phytologist © 2016 New Phytologist Trust.
Gene Expression dominance in allopolyploids: Hypotheses and models
DOI:S1360-1385(18)30014-1
PMID:29433919
[本文引用: 1]

The classical example of nonadditive contributions of the two parents to allopolyploids is nucleolar dominance, which entails silencing of one parental set of ribosomal RNA genes. This has been observed for many other loci. The prevailing explanation for this genome-wide expression disparity is that the two merged genomes differ in their transposable element (TE) complement and in their level of TE-mediated repression of gene expression. Alternatively, and not exclusively, gene expression dominance may arise from mismatches between trans effectors and their targets. Here, we explore quantitative models of regulatory mismatches leading to gene expression dominance. We also suggest that, when pairs of merged genomes are similar from one allopolyploidization event to another, gene-level and genome dominance patterns should also be similar.Copyright © 2018 Elsevier Ltd. All rights reserved.
Different patterns of ecological divergence between two tetraploids and their diploid counterpart in a parapatric linear coastal distribution polyploid complex
DOI:10.3389/fpls.2020.00315 URL [本文引用: 1]
Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants
Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell
DOI:10.1086/700636 URL [本文引用: 1]
Ancient whole genome duplications, novelty and diversification: The WGD Radiation Lag-Time Model
DOI:10.1016/j.pbi.2012.03.011
PMID:22480429
[本文引用: 1]

Many large and economically important plant groups (e.g. Brassicaceae, Poaceae, Asteraceae, Fabaceae and Solanaceae) have had ancient whole genome duplications (WGDs) occurring near or at the time of their origins, suggesting that WGD contributed to the origin of novel key traits and drove species diversification. However, these large clades show phylogenetic asymmetries with a species-rich crown group and a species-poor sister clade, suggesting significant 'lag-times' between WGDs and radiations. The species-poor sister groups share many key traits, but are often restricted to the hypothesized center of origin for the larger clade. Thus, the ultimate success of the crown group does not only involve the WGD and novel key traits, but largely subsequent evolutionary phenomena including later migration events, changing environmental conditions and/or differential extinction rates.Copyright © 2012 Elsevier Ltd. All rights reserved.
Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event
Angiosperms at the edge: Extremity, diversity, and phylogeny
Picking up the Ball at the K/Pg Boundary: The distribution of ancient polyploidies in the plant phylogenetic tree as a spandrel of asexuality with occasional sex
DOI:10.1105/tpc.16.00836 PMID:28213362 [本文引用: 2]
Evolution of Lomandroideae: Multiple origins of polyploidy and biome occupancy in Australia
DOI:S1055-7903(20)30108-1
PMID:32304826
[本文引用: 1]

Asparagaceae: Lomandroideae are a species-rich and economically important subfamily in the monocot order Asparagales, with a center of diversity in Australia. Lomandroideae are ecologically diverse, occupying mesic and arid biomes in Australia and possessing an array of key traits, including sexual dimorphism, storage organs and polyploidy that are potentially adaptive for survival in seasonally arid and fire-dependent habitats. The Lomandroideae phylogeny was reconstructed using maximum likelihood and Bayesian inference criteria, based on plastome data from genome-skimming to infer relationships. A fossil-calibrated chronogram provided a temporal framework for understanding trait transitions. Ancestral state reconstructions and phylogenetic comparative trait correlation analyses provided insights into the evolutionary and ecological drivers associated with Lomandroideae diversification. Lomandroideae diverged from the other Asparagaceae ca. 56.61 million years ago (95% highest posterior density values 70.31-45.34 million years) and the major lineages diversified since the Oligocene. The most recent common ancestor of the clade likely occupied the mesic biome, was hermaphroditic and geophytic. Biome occupancy transitions were correlated with polyploidy and the presence of storage roots. Polyploidy potentially serves as an "enabler" trait, generating novel phenotypes, which may confer tolerance to climatic ranges and soil conditions putatively required for expansion into and occupation of new arid biomes. Storage roots, as a key factor driving biome transitions, may have been associated with fire rather than with aridification events in the Australian flora. This study contributes significantly to our understanding of biome evolution by identifying polyploidy and storage organs as key factors associated with transitions in biome occupancy in this lineage.Copyright © 2020 Elsevier Inc. All rights reserved.
Effect of polyploidisation on the response of apple (Malus ×domestica Borkh.) toVenturia inaequalis infection
DOI:10.1007/s10658-017-1395-2 URL [本文引用: 1]
A time-calibrated road map of Brassicaceae species radiation and evolutionary history
DOI:10.1105/tpc.15.00482
PMID:26410304
[本文引用: 1]

The Brassicaceae include several major crop plants and numerous important model species in comparative evolutionary research such as Arabidopsis, Brassica, Boechera, Thellungiella, and Arabis species. As any evolutionary hypothesis needs to be placed in a temporal context, reliably dated major splits within the evolution of Brassicaceae are essential. We present a comprehensive time-calibrated framework with important divergence time estimates based on whole-chloroplast sequence data for 29 Brassicaceae species. Diversification of the Brassicaceae crown group started at the Eocene-to-Oligocene transition. Subsequent major evolutionary splits are dated to ∼20 million years ago, coinciding with the Oligocene-to-Miocene transition, with increasing drought and aridity and transient glaciation events. The age of the Arabidopsis thaliana crown group is 6 million years ago, at the Miocene and Pliocene border. The overall species richness of the family is well explained by high levels of neopolyploidy (43% in total), but this trend is neither directly associated with an increase in genome size nor is there a general lineage-specific constraint. Our results highlight polyploidization as an important source for generating new evolutionary lineages adapted to changing environments. We conclude that species radiation, paralleled by high levels of neopolyploidization, follows genome size decrease, stabilization, and genetic diploidization. © 2015 American Society of Plant Biologists. All rights reserved.
Ecological differentiation, lack of hybrids involving diploids, and asymmetric gene flow between polyploids in narrow contact zones ofSenecio carniolicus (syn. Jacobaea carniolica, Asteraceae)
DOI:10.1002/ece3.2015.5.issue-6 URL [本文引用: 1]
Ancestral polyploidy in seed plants and angiosperms
DOI:10.1038/nature09916 URL [本文引用: 1]
Asexuality and the coexistence of cytotypes
DOI:10.1111/nph.2007.175.issue-4 URL [本文引用: 1]
Mixed-ploidy species: Progress and opportunities in polyploid research
DOI:10.1016/j.tplants.2017.09.011 URL [本文引用: 8]
Minority cytotype exclusion in local plant populations
DOI:10.2307/1218997 URL [本文引用: 2]
Morphological and cytotype variation of wild kiwifruit (Actinidia chinensis complex) along an altitudinal and longitudinal gradient in central-west China
DOI:10.1111/boj.2010.164.issue-1 URL [本文引用: 1]
Integrative taxonomy resolved species delimitation in a fern complex: A case study of theAsplenium coenobiale complex
DOI:10.17520/biods.2019316
[本文引用: 1]

Due to wide hybridization and polyploidization, there are numerous species complexes with taxonomic challenges in the fern genus Asplenium. Integrative taxonomy using evidence of morphology, cytology and molecular phylogeny provides one of the best ways for the discovery and delimitation of species. The Asplenium coenobiale complex represents one of the spleenwort complexes, which are morphologically variable and difficult in species delimitation. Owing to the lack of comprehensive sampling and systematic study, the taxonomy of this complex remains unresolved. In this study, we selected representative individuals of this complex covering differences in morphology and geography. We conducted a palynological study to explore reproductive characteristics, and inferred the ploidy level through flow cytometry. Furthermore, based on the phylogenetic evidence from chloroplast and nuclear genomes, we discussed the evolutionary relationship and origin of this complex. Our results showed that: (1) The development of 64 spores within a single normal sporangium is indicative of the ability of sexual reproduction, although abortive sporangia are common in the Asplenium coenobiale complex. (2) Ploidy variation is found in this complex, i.e. A. cornutissimum is diploid, whereas other members are all tetraploid. (3) The maternally inherited chloroplast phylogeny supported four clades within this complex, and this was incongruent with the nuclear phylogeny; therefore, it was inferred that hybridization could be an important driving force during the formation of the complex. Based on our analyses, we conduct a revision to the A. coenobiale complex, i.e. one newly discovered autotetraploid species (A. maguanense sp. nov.), one diploid species (A. cornutissimum), and two allotetraploids with reciprocal origins (A. coenobiale and A. pulcherrimum).
利用整合分类学方法进行蕨类植物复合体的物种划分: 以线裂铁角蕨复合体为例
DOI:10.17520/biods.2019316
[本文引用: 1]

广泛的杂交和多倍化使得铁角蕨属(Asplenium)下存在着许多分类困难的物种复合体, 针对这些类群进行整合分类学的研究, 有助于我们更加全面和深入地理解物种的界限以及形成机制。线裂铁角蕨复合体(Asplenium coenobiale complex)是铁角蕨属下一个形态多样性较高的类群, 由于缺乏全面取样和系统研究, 该复合体的物种划分长期存在争议。本研究选取线裂铁角蕨复合体中形态变异和地理分布具有代表性的个体, 通过孢粉学研究确定该类群的生殖特性, 运用流式细胞分析获取倍性信息, 同时结合叶绿体和核基因组片段系统发生分析的证据, 对该类群的系统演化关系和起源方式进行了探讨。结果表明: (1)虽然部分孢子囊败育的情况在线裂铁角蕨复合体中十分普遍, 但正常孢子囊内形成的64个孢子说明该类群植物仍能进行正常的有性生殖; (2)该复合体中存在着倍性变异, 其中多角铁角蕨(A. cornutissimum)是二倍体, 而其他成员均为四倍体; (3)依据母系遗传的叶绿体序列所构建的系统发生关系将该类群划为4个分支, 与基于核基因序列构建的系统树存在冲突, 这暗示杂交可能在该复合体的形成过程中起到了重要的推动作用。综上所述, 我们建议将线裂铁角蕨复合体划分为4个物种, 即同源四倍体新种马关铁角蕨(A. maguanense sp. nov.), 二倍体多角铁角蕨, 以及两个由同一对亲本正反交产生的异源四倍体线裂铁角蕨(A. coenobiale)和叶基宽铁角蕨(A. pulcherrimum)。
Flow cytometry, microsatellites and niche models reveal the origins and geographical structure ofAlnus glutinosa populations in Europe
DOI:10.1093/aob/mcv158 URL [本文引用: 1]
Recently formed polyploid plants diversify at lower rates
DOI:10.1126/science.1207205
PMID:21852456
[本文引用: 2]

Polyploidy, the doubling of genomic content, is a widespread feature, especially among plants, yet its macroevolutionary impacts are contentious. Traditionally, polyploidy has been considered an evolutionary dead end, whereas recent genomic studies suggest that polyploidy has been a key driver of macroevolutionary success. We examined the consequences of polyploidy on the time scale of genera across a diverse set of vascular plants, encompassing hundreds of inferred polyploidization events. Likelihood-based analyses indicate that polyploids generally exhibit lower speciation rates and higher extinction rates than diploids, providing the first quantitative corroboration of the dead-end hypothesis. The increased speciation rates of diploids can, in part, be ascribed to their capacity to speciate via polyploidy. Only particularly fit lineages of polyploids may persist to enjoy longer-term evolutionary success.
Derived alleles of two axis proteins affect meiotic traits in autotetraploidArabidopsis arenosa
Anthropogenic disturbance as a driver of microspatial and microhabitat segregation of cytotypes ofCentaurea stoebe and cytotype interactions in secondary contact zones
DOI:10.1093/aob/mcs120 URL [本文引用: 1]
Selection for phenotypic divergence between diploid and autotetraploidHeuchera grossulariifolia
Much of the diversity of flowering plants is associated with genomic duplication through polyploidy. Little is known, however, about the evolutionary mechanisms responsible for the diversification of novel polyploid lineages. We evaluated the possibility that divergence is driven by natural selection by estimating the strength of phenotypic selection acting on three floral traits in sympatric populations of diploid and autotetraploid Heuchera grossulariifolia over three years. Our results demonstrate consistent directional selection for increasing scape length and floral display in both diploid and tetraploid populations. In contrast, selection acting on flowering phenology varied across year and ploidy. Specifically, selection was found to favor late-flowering diploids in 2001 and 2002 but early-flowering tetraploids in 2003. We investigated the mechanistic basis of divergent selection for flowering phenology in 2003 by estimating the relationship between plant flowering phenology and the probability of intercytotype pollinator movement. The results demonstrated that less divergent tetraploids were significantly more likely to experience intercytotype flights than were more divergent tetraploids. This result is consistent with the pattern of phenotypic selection observed. Taken together, our results suggest that divergence of polyploids and their diploid progenitors may be driven by a process analogous to reinforcement whereby selection favors phenotypes that reduce the probability of intercytotype matings with reduced fertility.
Ecological studies of polyploidy in the 100 years following its discovery
DOI:10.1098/rstb.2013.0352 URL [本文引用: 1]
Pathways, mechanisms, and rates of polyploid formation in flowering plants
DOI:10.1146/annurev.ecolsys.29.1.467 URL [本文引用: 1]
Analysis of ploidy segregation and genetic variation of progenies of different interploidy crosses inActinidia chinensis
中华猕猴桃不同倍性间杂交后代倍性分离和遗传变异分析
Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms
DOI:S1674-2052(18)30022-4
PMID:29317285
[本文引用: 2]

Gene duplications provide evolutionary potentials for generating novel functions, while polyploidization or whole genome duplication (WGD) doubles the chromosomes initially and results in hundreds to thousands of retained duplicates. WGDs are strongly supported by evidence commonly found in many species-rich lineages of eukaryotes, and thus are considered as a major driving force in species diversification. We performed comparative genomic and phylogenomic analyses of 59 public genomes/transcriptomes and 46 newly sequenced transcriptomes covering major lineages of angiosperms to detect large-scale gene duplication events by surveying tens of thousands of gene family trees. These analyses confirmed most of the previously reported WGDs and provided strong evidence for novel ones in many lineages. The detected WGDs supported a model of exponential gene loss during evolution with an estimated half-life of approximately 21.6 million years, and were correlated with both the emergence of lineages with high degrees of diversification and periods of global climate changes. The new datasets and analyses detected many novel WGDs widely spread during angiosperm evolution, uncovered preferential retention of gene functions in essential cellular metabolisms, and provided clues for the roles of WGD in promoting angiosperm radiation and enhancing their adaptation to environmental changes.Copyright © 2018 The Author. Published by Elsevier Inc. All rights reserved.
The Chromosome Counts Database (CCDB)—A community resource of plant chromosome numbers
DOI:10.1111/nph.2015.206.issue-1 URL [本文引用: 1]
The global biogeography of polyploid plants
Plant speciation
Like the formation of animal species, plant speciation is characterized by the evolution of barriers to genetic exchange between previously interbreeding populations. Prezygotic barriers, which impede mating or fertilization between species, typically contribute more to total reproductive isolation in plants than do postzygotic barriers, in which hybrid offspring are selected against. Adaptive divergence in response to ecological factors such as pollinators and habitat commonly drives the evolution of prezygotic barriers, but the evolutionary forces responsible for the development of intrinsic postzygotic barriers are virtually unknown and frequently result in polymorphism of incompatibility factors within species. Polyploid speciation, in which the entire genome is duplicated, is particularly frequent in plants, perhaps because polyploid plants often exhibit ecological differentiation, local dispersal, high fecundity, perennial life history, and self-fertilization or asexual reproduction. Finally, species richness in plants is correlated with many biological and geohistorical factors, most of which increase ecological opportunities.
Cytotype coexistence leads to triploid hybrid production in a diploid-tetraploid contact zone ofChamerion angustifolium (Onagraceae)
DOI:10.3732/ajb.1200583
PMID:23629844
[本文引用: 1]

Polyploids are often geographically segregated from their diploid progenitors, but the extent of sympatry and the consequences for reproductive isolation and coexistence are rarely quantified. •In this study, we document the distribution and co-occurrence of diploid and tetraploid Chamerion angustifolium among 57 populations within the diploid-tetraploid contact zone in the Canadian Rocky Mountains. Rates of hybrid mating in mixed-ploidy populations were inferred from the frequency of triploid offspring in open-pollinated seed families. •Twenty-three of 57 populations sampled contained a single cytotype; 20 (87%) were tetraploid and three (13%) were diploid. Thirty-four populations (60%) contained multiple ploidies. Diploid and tetraploid plants occurred in all mixed-ploidy populations; triploids occurred in 13 populations and averaged 1.4% of plants per population. The proportion of tetraploids in a population was negatively related to elevation (partial regression: F = 27.2, P <0.0001) and latitude (partial regression: F = 17.4, P < 0.0001). Triploids were detected in seed from all eight mixed-ploidy populations sampled ( = 3.7% of seed per population), comprising 7% of that expected with random mating (G = 2589.2, df = 1, P <0.0001, n = 2628), and were more often produced by diploid maternal parents than tetraploid parents. •Our results indicate that tetraploids regularly coexist with diploids in the contact zone and that this coexistence is likely promoted by both strong reproductive isolation and asymmetric intercytotype mating between diploid and tetraploid C. angustifolium.
Polyploidy and angiosperm diversification
DOI:10.3732/ajb.0800079 URL [本文引用: 1]
Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al (2011)
DOI:10.1111/nph.2014.202.issue-4 URL [本文引用: 3]
Ecological differentiation of diploid and polyploid cytotypes ofSenecio carniolicus sensu lato (Asteraceae) is stronger in areas of sympatry
DOI:10.1093/aob/mcv176
PMID:26658487
[本文引用: 1]

Ecological differentiation is recognized as an important factor for polyploid speciation, but little is known regarding whether the ecological niches of cytotypes differ between areas of sympatry and areas where single cytotypes occur (i.e. niche displacement).Ecological niches of four groups of Senecio carniolicus sensu lato (s.l.) (western and eastern diploid lineages, tetraploids and hexaploids) were characterized via Landolt indicator values of the accompanying vascular plant species and tested using multivariate and univariate statistics.The four groups of S. carniolicus s.l. were ecologically differentiated mainly with respect to temperature, light and soil (humus content, nutrients, moisture variability). Niche breadths did not differ significantly. In areas of sympatry hexaploids shifted towards sites with higher temperature, less light and higher soil humus content as compared with homoploid sites, whereas diploids and tetraploids shifted in the opposite direction. In heteroploid sites of tetraploids and the western diploid lineage the latter shifted towards sites with lower humus content but higher aeration.Niche displacement can facilitate the formation of stable contact zones upon secondary contact of polyploids and their lower-ploid ancestors and/or lead to convergence of the cytotypes' niches after they have attained non-overlapping ranges. Niche displacement is essential for understanding ecological consequences of polyploidy.© The Author 2015. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Polyploidy affects the seed, dormancy and seedling characteristics of a perennial grass, conferring an advantage in stressful climates
DOI:10.1111/plb.13094
PMID:32011086
[本文引用: 1]

Polyploidy (the state of having more than two genome copies) is widely distributed in flowering plants and can vary within species, with polyploid races often associated with broad ecological tolerances. Polyploidy may influence within-species variation in seed development, germination and establishment. We hypothesized that interactions between polyploidy and the seed developmental environment would affect subsequent dormancy, germination and early growth traits, particularly in stressful environments. Using seeds developed in a common garden under ambient and warmed conditions, we conducted germination trials under drought and temperature stress, and monitored the subsequent growth of seedlings. The study species, Themeda triandra, is a widespread, keystone, Australian native grass and a known polyploid complex. Tetraploid plants produced heavier, more viable seeds than diploids. Tetraploids were significantly more dormant than diploids, regardless of seed developmental environment. Non-dormant tetraploids were more sensitive to germination stress compared to non-dormant diploids. Finally, tetraploid seedlings were larger and grew faster than diploids, usually when maternal plants were exposed to developmental temperatures atypical to the source environment. Seed and seedling traits suggest tetraploids are generally better adapted to stressful environments than diploids. Because tetraploid seeds of T. triandra are more dormant they are less likely to germinate under stress, and when they do germinate, seedling growth is rapid and independent of seed developmental environment. These novel results demonstrate that polyploidy, sometimes in interaction with developmental environment and possibly also asexuality, can have within-species variation in seed and seedling traits that increase fitness in stressful environments.© 2020 German Society for Plant Sciences and The Royal Botanical Society of the Netherlands.
Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plantSenecio carniolicus (Asteraceae)
DOI:10.3732/ajb.94.8.1391
PMID:21636507
[本文引用: 2]

DNA ploidy levels were estimated using DAPI-flow cytometry of silica-dried specimens of the European mountain plant Senecio carniolicus (Asteraceae), covering its entire distribution area in the Eastern Alps (77 populations, 380 individuals) and the Carpathians (five populations, 22 individuals). A complex pattern of ploidy level variation (2x, 4x, 5x, 6x, and 7x cytotypes) was found in this species, which has been considered uniformly hexaploid. Hexaploids predominated in the Eastern Alps and was the only cytotype found in the Carpathians, while odd ploidy levels (5x, 7x) constituted a small fraction of the samples (<1.3%). Tetraploids occurred in two disjunct areas, which correspond with putative Pleistocene refugia for silicicolous alpine plants. Diploids occurred in large portions of the Alps but were absent from areas most extensively glaciated in the past. Intrapopulational cytotype mixture was detected in 22 populations-the majority involving diploids and hexaploids-with intermediate ploidy levels mostly lacking, suggesting limited gene flow and the evolution of reproductive isolation. Significant and reproducible intracytotype variation in nuclear DNA content was observed. Higher genome size in western diploids might be due to ancient introgression with the closely related S. incanus or to different evolutionary pathways in the geographically separated diploids.
Postzygotic isolation varies by ploidy level within a polyploid complex
DOI:10.1111/nph.14116
PMID:27533526
[本文引用: 1]

Whole genome duplication is considered to be a significant contributor to angiosperm speciation due to accumulation of rapid, strong interploid reproductive isolation. However, recent work suggests that interploid reproductive isolation may not be complete, especially among higher order cytotypes. This study evaluates postzygotic reproductive isolation among three cytotypes within a polyploid complex. We conducted reciprocal crosses using two diploid and two hexaploid populations each crossed to tetraploid populations spanning the geographic and phylogenetic range of the Campanula rotundifolia polyploid complex. Interploid and intrapopulation crosses were scored for fruit set, seed number, germination proportion and pollen viability. Postzygotic isolation was calculated for each cross as the product of these fitness components. A subset of offspring was cytotyped via flow cytometry. Postzygotic isolation was significantly lower in tetraploid-hexaploid crosses than diploid-tetraploid crosses, mostly due to substantially higher germination among tetraploid-hexaploid crosses. Tetraploid-hexaploid crosses produced pentaploids exclusively, whereas diploid-tetraploid crosses produced both triploids and tetraploids in high frequencies. Postzygotic isolation was weaker among higher order polyploids than between diploids and tetraploids, and unreduced gametes may facilitate diploid-tetraploid reproduction. This incomplete postzygotic isolation could allow ongoing interploid gene flow, especially among higher order polyploids, which may slow divergence and speciation in polyploid complexes.© 2016 The Authors. New Phytologist © 2016 New Phytologist Trust.
The genome of the mesopolyploid crop speciesBrassica rapa
DOI:10.1038/ng.919 URL [本文引用: 1]
Polyploidy: An evolutionary and ecological force in stressful times
DOI:10.1093/plcell/koaa015 URL [本文引用: 3]
The evolutionary significance of polyploidy
DOI:10.1038/nrg.2017.26 URL [本文引用: 5]
Induction of tetraploids inImpatiens (Impatiens walleriana) and characterization of their changes in morphology and resistance to downy mildew
DOI:10.21273/HORTSCI13093-18 URL [本文引用: 1]
The frequency of polyploid speciation in vascular plants
Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms
DOI:10.1016/j.molp.2019.10.012 URL [本文引用: 3]
Meiotic adaptation to genome duplication inArabidopsis arenosa
DOI:10.1016/j.cub.2013.08.059 URL [本文引用: 1]
Distribution pattern of ploidy variation ofActinidia chinensis andA. deliciosa
中华猕猴桃和美味猕猴桃的倍性变异及地理分布研究
Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation
Plant polyploidy: Origin, evolution, and its influence on crop domestication
DOI:10.1016/j.hpj.2019.11.003
[本文引用: 3]

The prevalence and recurrence of polyploidization in plant species make it one of the most important evolutionary events in plants, and as a result, polyploidization is an extensively investigated research field. Due to the rapid development of sequencing technologies, there is increased evidence to support that polyploidization plays an important role in the diversification of plant species, evolution of genes, and the domestication of crops. Here, we reviewed the influence of polyploidization on various aspects of plant evolution, mainly focused on polyploid origin, characteristics, subsequent genome divergence, and its impact on gene function innovation and crop domestication. The occurrence of many independent polyploidization events in plants was found to be tightly associated with the timing of extreme climate events or natural disasters on earth, leading to mass extinction while possibly facilitating increased polyploidization. Following allo-polyploidization, a distinct phenomenon known as sub-genome dominance occurred during sub-genome evolution, which was found to be associated with the methylation of transposons. Extensive gene fractionations (lost) following polyploidization were reported in almost all paleo-polyploids, and the evolutionary fates of multi-copy genes, such as sub-/neo- functionalization, were further proposed to illustrate their underlying mechanisms. Moreover, polyploidization was found to significantly impact species diversification, with subsequent effects on crop domestication and the development of traits with agronomic importance. Based on the progress of plant polyploidization studies, we discussed several main topics that might further improve our understanding of polyploid evolution and that are likely contribute to the application of polyploidization in crop breeding in the near future.