天然胡杨(Populus euphratica)是中国西北干旱荒漠区重要的珍稀乔木树种, 被列为世界珍稀濒危保护林木资源(王世绩等, 1995)。全世界61%的胡杨分布在中国, 而中国91%的胡杨林分布在新疆(王世绩等, 1995), 胡杨林对新疆沙漠地区的生态环境起到防风固沙的作用, 而且其耐盐碱、抗干旱等优异抗逆的特性(顾亚亚等, 2013)对未来培育其他适应于盐碱地环境的植物提供了理论上的可行性。近年来, 由于生境破坏和人类活动的增加, 胡杨林面积严重减少或退化, 加强对现有极端荒漠地区胡杨群体和优异种质资源的保护显得格外重要。一般而言, 原地保存和异地保存是植物群体保护的两种主要方式(Dzialuk et al, 2014; Nagamitsu et al, 2014)。原地保存通常针对群体间无明显的遗传分化, 且遗传多样性高的群体(Dzialuk et al, 2014; Tijerino & Korpelainen, 2014), 异地保存通常针对群体间存在分化且遗传多样性低的群体(李斌和顾万春, 2005)。胡杨种质资源分布范围广泛, 对整个分布区的资源进行保护难度较大。因此, 在有限条件下保护有价值或有代表性的种质资源, 不失为一种经济有效的保护策略。在此过程中, 还需要全面衡量不同胡杨群体分布区面积大小、遗传多样性水平高低、分布区生态环境、实施保护的难易程度和经济成本等因素(Sáenz-romero et al, 2003)。
前期胡杨群体遗传学研究局限于狭窄地理区域, 采用的分子标记、个体或群体数量相对有限(Wu et al, 2008; Eusemann et al, 2013; Xu et al, 2013; Wang et al, 2014), 或使用较为传统的非共显性标记(如AFLP、RAPD和SRAP) (Saito et al, 2002; Wang et al, 2011; Kansu & Kaya, 2020)。此外, 利用核标记和叶绿体DNA标记对中国西北地区胡杨群体结构和谱系地理学的初步研究表明, 新疆南疆和北疆胡杨群体存在明显的分化, 且南疆群体遗传多样性低于北疆(Zeng et al, 2018)。然而, 利用全基因组重测序对胡杨的27个天然群体252个个体的遗传多样性研究表明, 南疆和北疆群体间遗传分化程度较高,南疆群体的遗传多样性高于北疆(Jia et al, 2020)。近期谱系生物地理学研究表明, 胡杨群体的遗传多样性可能与地理距离和环境因子有关, 其中遗传多样性与经度和纬度呈显著负相关, 且纬度对遗传多样性的影响更大, 而与最暖季和最干月降水量、年平均气温等环境因子呈显著正相关(Gai et al, 2021)。先前的研究表明胡杨和灰杨(Populus pruinosa)存在着基因流和分化时间的差异, 进而促进了胡杨物种形成的复杂性(Ma et al, 2018)。因此, 研究胡杨的遗传多样性和群体结构, 对于探索其适应性进化以及群体间的遗传关系具有重要意义。
单核苷酸多态性(single nucleotide polymorphisms, SNPs)指单个核苷酸变异, 包括碱基转换、颠换等。SNPs具有二态性、高频率和稳定遗传等特性, 被认为是继限制性片段多态性标记、微卫星标记之后的第三代DNA遗传标记。随着高通量测序技术的发展, SNPs标记的开发也变得相对容易, 因此被广泛应用于人类、动植物的分子遗传学研究中, 如基因的关联定位、遗传图谱的构建及群体遗传多样性的分析等(Kaiser et al, 2017)。
1 材料与方法
1.1 研究对象
2020年通过对中国西北地区天然胡杨林的实地考察, 获得了58个群体的采样点信息(表1)。其中每个群体选取30株左右的成年样株, 从每样株上取当年生条上基部开始第3节位的叶片, 在野外用硅胶干燥保存用于总DNA提取。为了减少无性系分株的影响, 每个群体所有取样个体至少间隔100 m, 同时对SNP基因型完全一致的个体进行去冗余。
表1 本研究中58个胡杨群体采样信息
Table 1
| 地区 Region | 样点 Site | 群体 Population | 经纬度 Location | 海拔 Altitude (m) | ||
|---|---|---|---|---|---|---|
| 南疆阿克苏 Aksu, SX | 柯坪县 Kalpin County | AKS-KPX | 79.52° E, 40.22° N | 1,042-1,095 | ||
| 阿瓦提县 Awat County | AKS-AWT | 80.94° E, 40.36° N | 1,000-1,036 | |||
| 温宿县 Wensu County | AKS-WSX | 80.79° E, 41.21° N | 1,104-1,140 | |||
| 沙雅县 Shaya County | AKS-SYX | 83.42° E, 41.59° N | 929-989 | |||
| 库车县 Kuqa County | AKS-KCS | 83.86° E, 41.28° N | 917-934 | |||
| 拜城县 Baicheng County | AKS-BCX | 81.81° E, 41.66° N | 1,231-1,256 | |||
| 阿拉尔市 Alaer City | AKS-ALE | 81.03° E, 40.51° N | 1,012-1,038 | |||
| 南疆喀什 Kashgar, SX | 巴楚县 Marabishi County | KS-BCX | 78.31° E, 39.77° N | 1,121-1,137 | ||
| 麦盖提县 Makit County | KS-MGTX | 78.24° E, 39.36° N | 1,132-1,156 | |||
| 岳普湖县 Yopurga County | KS-YPHX | 77.24° E, 39.18° N | 1,166-1,246 | |||
| 伽师县 Payzawat County | KS-JSX | 77.38° E, 39.65° N | 1,161-1,174 | |||
| 疏勒县 Shule County | KS-SLX | 76.18° E, 30.17° N | 1,259-1,279 | |||
| 莎车县 Yarkant County | KS-SCX | 77.36° E, 38.40° N | 1,187-1,220 | |||
| 叶城县 Yecheng County | KS-YCX | 77.58° E, 38.18° N | 1,230-1,299 | |||
| 南疆和田 Hotan, SX | 皮山县 Pishan County | HT-PSX | 79.01° E, 37.62° N | 1,290-1,328 | ||
| 墨玉县 Karakax County | HT-MYX | 90.19° E, 37.77° N | 1,239-1,290 | |||
| 洛浦县 Lop County | HT-LPX | 80.12° E, 37.43° N | 1,101-1,284 | |||
| 策勒县 Qira County | HT-CLX | 81.03° E, 36.94° N | 1,394-1,457 | |||
| 于田县 Yutian County | HT-YTX | 81.41° E, 37.60° N | 1,263-1,452 | |||
| 民丰县 Minfeng County | HT-MFX | 82.79° E, 37.73° N | 1,278-1,387 | |||
| 南疆巴音郭楞蒙古自治州 Bayingol Mongolian Autonomous Prefecture, SX | 且末县 Qiemo County | BZ-QMX | 84.09° E, 37.73° N | 1,088-1,315 | ||
| 若羌县 Ruoqiang County | BaZ-RQX | 87.12° E, 38.78° N | 801-984 | |||
| 尉犁县 Yuli County | BaZ-YLX | 85.77° E, 41.14° N | 841-931 | |||
| 库尔勒市 Korla City | BaZ-KEL | 85.78° E, 41.53° N | 884-895 | |||
| 和硕县 Heshuo City | BaZ-HSX | 86.85° E, 42.17° N | 1,047-1,084 | |||
| 轮台县 Bugur County | BaZ-LTX | 84.25° E, 41.26° N | 905-969 | |||
| 甘肃 Gansu | 民勤县 Minqin County | GS-MQX | 103.1° E, 38.65° N | 1,283-1,323 | ||
| 金塔县 Jinta County | GS-JTX | 98.87° E, 39.93° N | 1,191-1,284 | |||
| 瓜州县 Guazhou County | GS-GZX | 96.05° E, 40.54° N | 1,027-1,218 | |||
| 敦煌县 Dunhuang County | GS-DHX | 94.83° E, 40.17° N | 1,033-1,140 | |||
| 阿克塞哈萨克族自治县 Kazak Autonomous County of Aksay | GS-AKS | 93.43° E, 39.78° N | 1,447-1,493 | |||
| 宁夏 Ningxia | 中卫市 Zhongwei City | NIX-ZW | 105.12° E, 37.59° N | 1,207-1,220 | ||
| 内蒙古 Inner Mongolia | 额济纳旗 Ejina | NMG-EJN | 101.08° E, 41.97° N | 813-926 | ||
| 四子王旗 Siziwang Banner | NMG-SZWQ | 111.27° E, 42.83° N | 901-963 | |||
| 青海 Qinghai | 格尔木市 Geermu City | QH-GEM | 94.33° E, 36.44° N | 2,709-2,936 | ||
| 北疆吐鲁番 Turpan, NX | 托克逊县 Tuokexun County | TLF-TKX | 88.17° E, 43.21° N | 1,128-1,152 | ||
| 北疆哈密 Hami, NX | 伊州区 Yizhou District | HM-YZQ | 93.08° E, 42.59° N | 390-405 | ||
| 伊吾县 Yiwu County | HM-YWX | 95.25° E, 43.66° N | 456-489 | |||
| 巴里坤县 Barkol County | HM-BLKX | 93.97° E, 43.97° N | 1,049-1,120 | |||
| 北疆昌吉 Changji, NX | 木垒哈萨克自治县 Mulei Kazak Autonomous County | CJ-MLX | 91.29° E, 44.82° N | 730-756 | ||
| 吉木萨尔县 Jimusaer County | CJ-JMSE | 89.01° E, 44.07° N | 670-676 | |||
| 呼图壁县 Hutubi County | CJ-HTBX | 86.90° E, 44.52° N | 359-401 | |||
| 玛纳斯县 Manasi County | CJ-MNSX | 86.87° E, 44.66° N | 353-362 | |||
| 北疆阿勒泰 Altay, NX | 哈巴河县 Habahe County | ALT-HBH | 86.26° E, 47.92° N | 431-456 | ||
| 福海县 Fuhai County | ALT-FHX | 87.78° E, 47.21° N | 489-494 | |||
| 布尔津县 Burqin County | ALT-BEJ | 86.94° E, 47.66° N | 466-494 | |||
| 富蕴县 Fuyun County | ALT-FYX | 88.75° E, 46.91° N | 677-724 | |||
| 北疆博州 Bortala Mongol Autonomous Prefecture, NX | 阿拉山口市 Alashankou City | BoZ-ALSK | 82.97° E, 45.01° N | 194-206 | ||
| 精河县 Jinghe County | BoZ-JHX | 83.43° E, 44.91° N | 203-217 | |||
| 北疆塔城 Tarbagatay, NX | 沙湾县 Shawan County | TC-SWX | 85.77° E, 44.88° N | 312-328 | ||
| 乌苏市 Wusu City | TC-WUSU | 83.83° E, 44.80° N | 260-268 | |||
| 和丰县 Hefeng County | TC-HFX | 85.27° E, 46.26° N | 469-477 | |||
| 托里县 Toli County | TC-TLX | 85.32° E, 46.15° N | 426-431 | |||
| 北疆伊犁哈萨克自治州 Ili Kazak Autonomous Prefecture, NX | 霍城县 Huocheng County | YL-HCX | 80.55° E, 44.09° N | 631-638 | ||
| 察布查尔锡伯自治县 Qapqal Xibe Autonomous County | YL-CX | 80.69° E, 43.84° N | 528-542 | |||
| 北疆克拉玛依 Karamay, NX | 克拉玛依区 Karamay District | KLMYS | 85.14° E, 45.33° N | 271-278 | ||
| 乌尔禾区 Urho District | KLMY-WEH | 85.67° E, 46.13° N | 296-302 | |||
| 北疆乌鲁木齐 Urumqi, NX | 乌鲁木齐市 Urumqi City | WLMQ | 88.35° E, 43.28° N | 936-969 | ||
SX: 南疆; NX: 北疆; QH: 青海; GS: 甘肃; NIX: 宁夏; NM: 内蒙古。
SX, Southern Xinjiang; NX, Northern Xinjiang; QH, Qinghai; GS, Gansu; NIX, Ningxia; NM, Inner Mongolia.
1.2 天然胡杨群体遗传多样性分析
采用靶向测序技术(target sequencing)对样品进行基因型分析。首先基于公开发表的胡杨重测序数据(Jia et al, 2020)和本实验室的参考基因组数据, 进行高质量SNP检测, 从中选择胡杨19条染色体上均匀分布的120个区间作为靶向测序的目标区间。筛选标准如下: (1) SNP间隔大于50 kb; (2) SNP对应的最小等位基因频率(minor allele frequency)不小于0.4; (3)杂合率不超过55%; (4)基因型缺失率不超过25%, 同时也检测了获得的SNPs标记符合随机抽取的变异位点。利用改良的CTAB法(Allen et al, 2006)提取叶片的DNA, 用Qubit 3.0 (Agiannitopoulos et al, 2020)检测DNA纯度(Thermo Qubit 3.0), 再通过多重PCR捕获目标区间, 最后利用Illumina 2500平台进行高通量测序。得到测序数据之后, 首先通过BWA软件将其比对到胡杨参考基因组(
1.3 胡杨遗传多样性保护单元的构建及评价
通过计算不同胡杨天然群体的Nei’s相似性系数(Nei, 1972), 采用逐步聚类优先取样法(曾宪君等, 2014; Wang et al, 2021)。按照遗传相似性从大到小的顺序将群体进行排序, 其中在遗传相似性最大的2个群体中, 剔除遗传多样性低的群体, 保留的57个群体记为CU57, 剔除的群体标记为R1; 如果2个群体的遗传多样性水平比较接近, 则考虑这2个群体的分布地环境及其保护成本高低, 从而做出可行性的删减; 依此类推, 直到天然群体的数目到2为止。然后对逐步聚类获得的不同天然群体的保护单元与初始群体间进行显著性分析。从CU57到CU2进行分析, 直到差异显著时为止; 此时上一组合所构成的胡杨保护单元, 则满足以最小的群体量尽可能多地代表整个群体遗传多样性的要求(李慧峰等, 2013)。
1.4 胡杨古树遗传多样性保护单元的评价
胡杨古树是指估测树龄在100年以上的个体。利用卷尺测量每棵树的胸径, 然后通过Logistic方程表达式A = 13.679/(1+3.3476×exp (-0.2099 D))来估测树龄, 其中A是树龄, D是胸径(顾亚亚等, 2013)。本研究中对各群体胡杨古树单独统计, 计算其遗传多样性。
2 结果
2.1 胡杨群体结构及遗传多样性
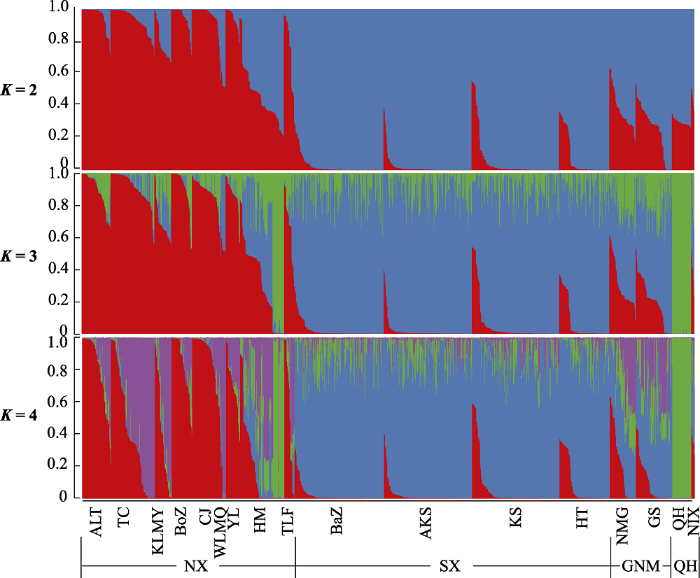
图1
图1
中国西北地区胡杨群体的群体结构。SX: 南疆; NX: 北疆; QH: 青海; GNM: 甘肃、宁夏和内蒙古。群体代号见
Fig. 1
The structural analysis of Populus euphratica populations in China. SX, Southern Xinjiang; NX, Northern Xinjiang; QH, Qinghai; GNM, Gansu, Ningxia, and Inner Mongolia. Population codes see
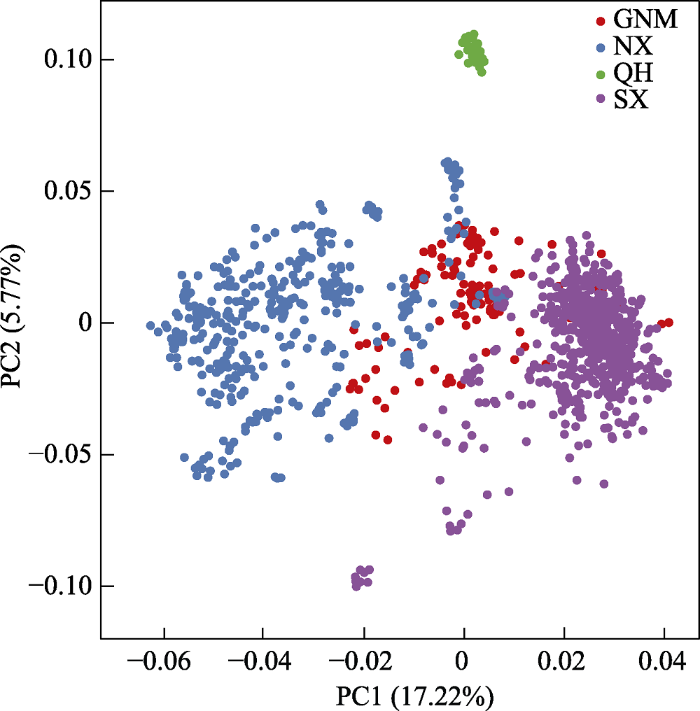
图2
图2
中国西北地区胡杨群体的主成分分析。SX: 南疆; NX: 北疆; QH: 青海; GNM: 甘肃、宁夏和内蒙古。
Fig. 2
The PCA analysis of Populus euphratica populations in China. SX, Southern Xinjiang; NX, Northern Xinjiang; QH, Qinghai; GNM, Gansu, Ningxia, and Inner Mongolia.
表2 58个天然胡杨群体的遗传多样性。群体代号同表1。
Table 2
| 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC | 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC |
|---|---|---|---|---|---|---|---|---|---|
| KS-MGTX | 0.294 | 0.262 | 0.443 | 0.235 | BoZ-JHX | 0.217 | 0.217 | 0.327 | 0.174 |
| AKS-ALE | 0.278 | 0.270 | 0.420 | 0.223 | TLF-TKX | 0.213 | 0.230 | 0.321 | 0.171 |
| KS-YPHX | 0.266 | 0.261 | 0.406 | 0.215 | AKS-WSX | 0.211 | 0.238 | 0.319 | 0.169 |
| HT-LPX | 0.261 | 0.264 | 0.390 | 0.208 | GS-DHX | 0.210 | 0.242 | 0.310 | 0.165 |
| HT-PSX | 0.248 | 0.255 | 0.378 | 0.200 | KLMYS | 0.210 | 0.211 | 0.315 | 0.167 |
| HT-YTX | 0.246 | 0.244 | 0.373 | 0.198 | TC-WUSU | 0.208 | 0.239 | 0.314 | 0.167 |
| KS-JSX | 0.246 | 0.243 | 0.378 | 0.199 | ALT-BEJ | 0.205 | 0.219 | 0.308 | 0.164 |
| NMG-EJN | 0.244 | 0.238 | 0.368 | 0.196 | TC-SWX | 0.204 | 0.229 | 0.306 | 0.164 |
| KS-BCX | 0.244 | 0.245 | 0.374 | 0.197 | HT-CLX | 0.203 | 0.258 | 0.304 | 0.161 |
| HT-MFX | 0.241 | 0.259 | 0.368 | 0.195 | GS-AKS | 0.200 | 0.249 | 0.303 | 0.161 |
| KS-SLX | 0.240 | 0.248 | 0.365 | 0.193 | CJ-HTB | 0.198 | 0.255 | 0.297 | 0.159 |
| NX-ZW | 0.236 | 0.261 | 0.351 | 0.188 | YL-HCX | 0.184 | 0.239 | 0.274 | 0.147 |
| AKS-SYX | 0.234 | 0.233 | 0.358 | 0.189 | CJ-JMSE | 0.177 | 0.199 | 0.266 | 0.142 |
| BaZ-KEL | 0.234 | 0.253 | 0.355 | 0.188 | YL-CX | 0.177 | 0.222 | 0.271 | 0.145 |
| GS-GZX | 0.231 | 0.245 | 0.347 | 0.185 | HM-BLKX | 0.176 | 0.225 | 0.255 | 0.137 |
| HT-MYX | 0.230 | 0.241 | 0.350 | 0.185 | TC-HFX | 0.174 | 0.221 | 0.261 | 0.139 |
| BaZ-HSX | 0.230 | 0.250 | 0.344 | 0.183 | ALT-FHX | 0.174 | 0.214 | 0.266 | 0.142 |
| KS-SCX | 0.228 | 0.246 | 0.347 | 0.184 | ALT-HBH | 0.174 | 0.210 | 0.267 | 0.142 |
| BaZ-RQX | 0.228 | 0.244 | 0.347 | 0.183 | KLMY-WEH | 0.172 | 0.245 | 0.259 | 0.137 |
| AKS-AWT | 0.228 | 0.237 | 0.351 | 0.185 | BoZ-ALSK | 0.167 | 0.208 | 0.262 | 0.138 |
| GS-JTX | 0.228 | 0.243 | 0.342 | 0.182 | HM-YZQ | 0.164 | 0.239 | 0.247 | 0.131 |
| BaZ-LTX | 0.226 | 0.232 | 0.340 | 0.180 | AKT-FYX | 0.154 | 0.239 | 0.225 | 0.120 |
| AKS-KCX | 0.225 | 0.228 | 0.340 | 0.181 | GS-MQX | 0.145 | 0.274 | 0.205 | 0.110 |
| AKS-BCX | 0.224 | 0.237 | 0.345 | 0.181 | CJ-MLX | 0.142 | 0.253 | 0.204 | 0.109 |
| CJ-MNS | 0.224 | 0.254 | 0.333 | 0.178 | WLMQ | 0.140 | 0.218 | 0.214 | 0.112 |
| BaZ-YLX | 0.222 | 0.245 | 0.339 | 0.179 | NMG-SZWQ | 0.128 | 0.237 | 0.182 | 0.098 |
| KS-YCX | 0.222 | 0.247 | 0.335 | 0.178 | QH-GEM | 0.126 | 0.233 | 0.181 | 0.097 |
| AKS-KPX | 0.221 | 0.243 | 0.333 | 0.177 | TC-TLX | 0.109 | 0.204 | 0.153 | 0.082 |
| BaZ-QMX | 0.218 | 0.241 | 0.329 | 0.175 | 平均 Mean | 0.208 | 0.239 | 0.314 | 0.167 |
| HM-YWX | 0.217 | 0.238 | 0.328 | 0.174 | 标准差 SD | 0.039 | 0.016 | 0.061 | 0.032 |
表3 胡杨群体内和群体间遗传变异的分子方差分析
Table 3
| 来源 Source | 自由度 Degree of freedom (df) | 平方和 Sum of square (SS) | 均方 Mean square (MS) | 方差分量 Variance component | 方差分量占比 Proportion (%) |
|---|---|---|---|---|---|
| 群体间 Among populations | 57 | 93,766.13 | 1,645.02 | 36.40 | 17.88 |
| 个体间 Among individuals | 1,103 | 215,156.92 | 195.07 | 27.96 | 13.74 |
| 个体内 Within individual | 1,161 | 161,558 | 139.15 | 139.15 | 68.38 |
| 总计 Total | 2,321 | 470,481.05 | - | 203.51 | 100 |
2.2 胡杨保护单元群体与初始群体间的差异性
根据不同群体的Nei’s相似性系数, 采用逐步聚类优先取样法将58个天然群体逐步分为56个保护单元(CU2-CU57), 分别与初始58个群体进行遗传多样性比较(附录2), 发现在不同抽样比例下各保护单元的标准差数值波动范围小, 且各个遗传多样性保护单元均与初始群体无显著差异。
通过胡杨保护单元群体(CU2-CU57)分别与58初始58个群体遗传多样性指标的显著性分析, 可以发现在期望杂合度(He)的t检验中, 保护单元(CU2-CU7、CU33-CU57)均与初始群体无显著差异(P > 0.05), 在观测杂合度(Ho)的t检验中, 保护单元(CU3-CU57)与初始群体均无显著差异, 在Shannon’s信息指数(I)和多态信息含量(PIC)的t检验中, 保护单元(CU2-CU4、CU7、CU33-CU57)与初始群体均无显著差异(附录3)。综合以上4个遗传多样性指标, 符合与初始群体相比无显著差异的胡杨保护单元群体分别为CU3、CU4、CU7、CU33-CU57, 其中保护单元最小群体为CU3, CU3的保留率分别为90.471% (He)、107.118% (Ho)、88.090% (I)、88.551% (PIC), 保护单元CU3的3个群体分别为甘肃省民勤群体(GS-MQX)、喀什地区麦盖提群体(KS-MGTX)和青海省格尔木群体(QH- GEM)。为了最大限度保护有代表性的胡杨群体, 构建的二级核心保护单元也应该能较好地代表初始群体的遗传多样性, 且与初始群体无显著差异。结果表明最适宜的二级保护单元群体为CU33, 保留率分别为92.376% (He)、99.668% (Ho)、91.872% (I)、92.046% (PIC)、He (92.376%)、Ho (99.668%)、I (91.872%)、PIC (92.046%)。
2.3 天然胡杨保护群体与剩余群体间的差异性
首先比较了保护单元与初始群体遗传多样性指标的差异显著性, 发现与初始群体相比均无显著差异的胡杨保护单元群体为CU3、CU4、CU7、CU33-CU57; 其次比较了胡杨保护单元群体与剩余群体遗传多样性指标的差异显著性, 发现在期望杂合度(He)、Shannon’s信息指数(I)和多态信息含量(PIC)的t检验中, 保护单元CU3、CU4、CU56均与初始群体无显著差异(P > 0.05); 在观测杂合度(Ho)的t检验中, 保护单元群体CU3、CU4、CU7、CU33-CU56与初始群体均无显著差异(附录4)。综合以上4个遗传多样性指标, 符合与剩余群体相比无显著差异的胡杨保护单元群体为CU3、CU4、CU56, 其中构建的保护单元最小群体为CU3, CU3的保留率分别为90.024% (He)、107.530% (Ho)、87.521% (I)、78.002% (PIC)。
2.4 遗传多样性保护单元的聚类分析
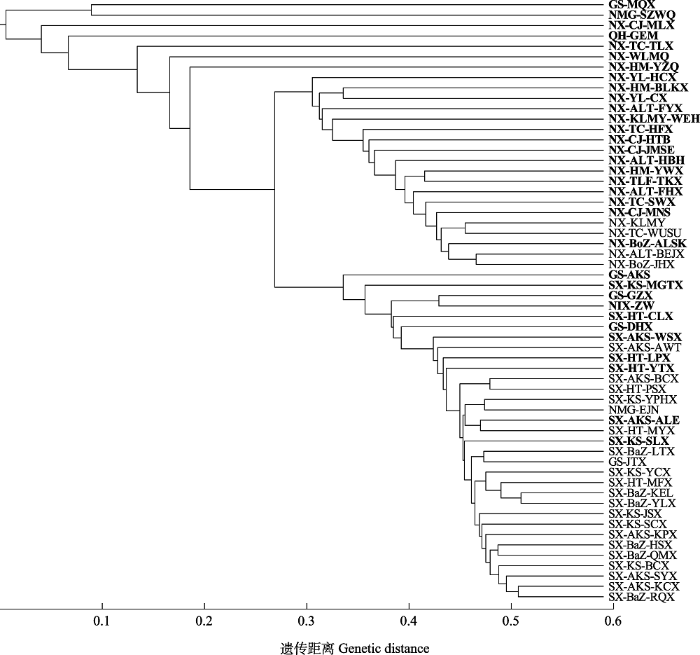
58个天然胡杨保护单元群体遗传距离的聚类结果(图3)表明, 当遗传距离等于0.3时, 58个群体分为两类, 且构建的33个保护单元群体也分为两类: 以南疆群体为代表聚为一类, 以北疆群体为代表聚为另一类, 进一步表明南疆和北疆的胡杨核心保护单元群体的遗传分化程度有显著差异。
图3
图3
58个胡杨群体遗传距离的聚类分析。群体采样信息见
Fig. 3
The cluster analysis based on genetic distance of 58 Populus euphratica populations. See
2.5 胡杨古树遗传多样性评价
23个胡杨古树群体的遗传多样性分析表明(表4), 古树群体的期望杂合度在0.144-0.274之间, 其中喀什地区麦盖提县古树群体(KS-MGTX)的期望杂合度最高, 昌吉地区木垒县古树群体(CJ-MLX)最低。Shannon’s信息指数(I)也表现出相同的趋势。结合胡杨地理分布区域综合分析表明, 胡杨古树主要分布在南疆, 且南疆地区群体的遗传多样性整体上高于北疆。
表4 23个胡杨古树群体的遗传多样性。群体代号同表1。
Table 4
| 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC | 群体 Population | 期望杂合度 He | 观测杂合度 Ho | Shannon’s 信息指数 I | 多态信息 含量 PIC |
|---|---|---|---|---|---|---|---|---|---|
| KS-MGTX | 0.274 | 0.268 | 0.439 | 0.236 | AKS-WSX | 0.221 | 0.256 | 0.331 | 0.176 |
| KS-BCX | 0.257 | 0.265 | 0.390 | 0.207 | BaZ-YLX | 0.218 | 0.247 | 0.326 | 0.174 |
| KS-YPHX | 0.252 | 0.277 | 0.377 | 0.201 | BaZ-QMX | 0.217 | 0.254 | 0.325 | 0.173 |
| AKS-SYX | 0.246 | 0.249 | 0.376 | 0.199 | BoZ-JHX | 0.212 | 0.231 | 0.308 | 0.164 |
| AKS-BCX | 0.244 | 0.250 | 0.373 | 0.197 | HT-MFX | 0.200 | 0.266 | 0.303 | 0.161 |
| HM-YWX | 0.236 | 0.258 | 0.354 | 0.189 | TC-WUSU | 0.198 | 0.240 | 0.286 | 0.152 |
| BaZ-RQX | 0.233 | 0.248 | 0.354 | 0.187 | BaZ-HSX | 0.181 | 0.259 | 0.261 | 0.140 |
| HT-PSX | 0.231 | 0.263 | 0.351 | 0.186 | TC-HFX | 0.175 | 0.229 | 0.262 | 0.139 |
| BaZ-KEL | 0.230 | 0.255 | 0.344 | 0.183 | KLMY-WEH | 0.167 | 0.258 | 0.253 | 0.134 |
| AKS-KCX | 0.225 | 0.243 | 0.339 | 0.180 | BoZ-ALSK | 0.164 | 0.221 | 0.251 | 0.133 |
| AKS-KPX | 0.224 | 0.267 | 0.329 | 0.176 | CJ-MLX | 0.144 | 0.260 | 0.211 | 0.113 |
| BaZ-LTX | 0.223 | 0.243 | 0.330 | 0.176 |
3 讨论
3.1 天然胡杨群体遗传多样性的分布格局
了解不同分布区天然胡杨群体遗传多样性水平有助于从分子水平上保护其遗传种质资源和构建核心种质库。本研究对采集的58个胡杨天然群体统计表明, 南疆群体最多(26个), 北疆次之(23个), 甘肃、青海、宁夏和内蒙古分布的群体最少。其中南疆群体的遗传多样性水平高于北疆及疆外群体(甘肃、青海、宁夏和内蒙古)。不同物种遗传多样性水平的高低与其分布区生态环境、物种繁殖能力及传播媒介等相关, 例如具有分布广泛、种子离体寿命长、风媒传播等生物学特性的物种相对较易克服群体间的地理隔离, 有利于不同群体间个体的杂交, 从而使群体维持较高水平的遗传多样性(Hamrick et al, 1979)。胡杨具有寿命长、根蘖繁殖、风媒传粉、种子借风传播等生物学特性, 有利于产生丰富的遗传变异, 从而适应不同的环境条件, 这可能是导致其群体遗传多样性丰富的主要原因(Wang et al, 2011)。同时, 南疆地区胡杨种子成熟常常发生于洪水期(Eusemann et al, 2013), 独特的环境条件有利于胡杨群体繁殖并建成大规模的有效群体, 使得南疆成为全球胡杨分布最密集的地区(王世绩等, 1995)。此外, 南疆地区有大量的灰杨分布, 胡杨和灰杨为雌雄异株的异交树种, 两物种开花阶段一致、花期重叠(王世绩等, 1995), 风媒传粉为种间基因交流提供了可能(Wang et al, 2011; 张玲等, 2012; Ma et al, 2018)。先前的研究也表明胡杨和灰杨之间有明显的基因渐渗现象(Wang et al, 2014; Jia et al, 2020)。这些可能是南疆地区胡杨群体具有较高遗传多样性和较低群体分化的主要原因。
3.2 胡杨遗传多样性保护单元的构建
保护一个物种全部种质资源的遗传多样性既困难又昂贵, 因此应该构建具有最小重复性和最大遗传多样性的核心子集, 以代表一个物种的整个基因库(Anoumaa et al, 2017)。在本研究中, 利用4个遗传多样性指标, 将胡杨保护单元群体分别与初始群体和剩余群体进行显著性分析, 得到与初始群体和剩余群体均无显著性差异的胡杨核心保护单元CU3、CU34和CU56。其中CU3样本量最小, 保留率最优, 因此定义CU3为一级胡杨核心保护单元群体(CU3群体为GS-MQX、KS-MGTX、QH-GEM)。其抽样比例为5.17%, 符合一般以5%-30%的抽样群体比例来覆盖整个初始群体的遗传变异(Wang et al, 2021)。考虑到胡杨的生态价值, 例如作为极端干旱区仅存乔木树种, 能够起到防风固沙、减缓土壤沙漠化的作用, 为了让核心保护群体不仅从遗传多样性水平同时能够从表型性状多样性方面代表整个群体的多样性水平, 选择原始58个群体大约一半的样本量作为胡杨的二级保护群体, 且保护群体需满足与原始群体无显著差异, 即此时二级保护单元群体也能较好地代表初始群体的遗传多样性。本文结果表明, 样本量最适宜的胡杨保护单元群体为CU33。定义CU33作为胡杨二级核心保护单元群体, 同时对CU33进行了聚类分析(图3), 结果表明当遗传距离等于0.3时, 新疆南疆和北疆的天然群体间出现了明显的遗传分化。
3.3 胡杨优先保护群体遗传多样性评价
同一物种不同群体的遗传多样性水平越高, 所构建的核心种质数目越大(曾宪君等, 2014)。然而在天然胡杨群体二级核心保护单元群体CU33中, 南疆大部分群体遗传多样性大于北疆群体(表2), 但南疆的保护单元群体数目(7个)小于北疆(19个) (图3)。可能的原因是南疆不同群体的Nei’s遗传相似性系数高, 意味着南疆群体内遗传多样性水平高、群体间遗传分化程度低; 其次由于生态环境的差异造成胡杨片断化的分布, 且北疆地区胡杨群体长期地理隔离, 导致群体间基因流动较少, 从而形成了群体内低的遗传多样性和群体间高的遗传分化(Zeng et al, 2018; Jia et al, 2020)。因此, 58个初始群体应选择更多的北疆群体作为保护单元群体, 在构建的二级核心保护单元群体CU33中, 北疆胡杨群体(19个)占比超过50%。保护单元CU33与初始58个群体在4个遗传多样性指标上的t检验均无显著差异, 更说明CU33能很好地代表初始58个群体。
天然胡杨群体拥有大量优异抗逆基因资源, 尤其对于古树资源的保护, 无论是对遗传多样性保护还是优异抗逆基因资源挖掘均具有重要的意义。在本研究中23个胡杨古树群体主要分布在南疆, 且南疆地区胡杨古树群体的遗传多样性整体上高于北疆。新疆位于干旱和半干旱区, 不同分布区水资源供需矛盾大, 干旱性气候频繁, 尤其在极端和严重干旱等级中, 南疆的年均干旱分布区大于北疆(张喜成等, 2020)。同时天山山脉阻隔了南北疆之间直接的基因流动, 造成了南北疆不同群体的遗传差异(Zeng et al, 2018; Jia et al, 2020)。南、北疆巨大的地理和环境差异是造成遗传天然胡杨群体遗传多样性分布不均一的重要原因。基于保护群体遗传相似性, 北疆的胡杨群体被优先作为保护单元; 同时北疆的胡杨遗传多样性低暗示着北疆的群体可能是经历天然选择的结果, 对现存的生态环境具有更好的适应性。然而, 南疆地区胡杨群体具有比北疆更丰富的遗传多样性和胡杨古树资源, 且南疆干旱程度较北疆严重。因此, 胡杨多样性的保护既要考虑遗传多样性丰富的群体, 确保为群体的扩大提供丰富的基因库; 也需要考虑经历自然选择而保留下的低遗传多样性群体, 有助于挖掘适应特定自然生态环境的优良基因型。综合考虑, 为了更好地保护胡杨种质资源, 核心保护单元群体的构建对于南北疆的胡杨资源的保护十分必要。
附录 Supplementary Material
附录1 胡杨4个亚群间遗传多样性比较
Appendix 1 Comparison of genetic diversity among four subpopulations
附录2 不同胡杨保护单元遗传多样性指标的多重比较
Appendix 2 Multiple comparison of genetic diversity indexes of different Populus euphratica conservation units
附录3 胡杨保护单元与初始群体期望杂合度(He)、观测杂合度(Ho)、Shannon’s信息指数(I)、多态信息含量(PIC)的t检验
Appendix 3 The t-test of the expected heterozygosity (He), observed heterozygosity (Ho), Shannon’s information index (I), polymorphism information content (PIC) between conservation units and all populations of Populus euphratica
附录4 胡杨核心保护单元与剩余群体期望杂合度(He)、观测杂合度(Ho)、Shannon’s信息指数(I)、多态信息含量(PIC)的t检验
Appendix 4 The t-test of the expected heterozygosity (He), observed heterozygosity (Ho), Shannon’s information index (I), polymorphism information content (PIC) between the conservation units and the rest populations of Populus euphratica
致谢
感谢张山河、孙健皓、韩晓莉、李秀、王佳宁、董晓山和王子健等同学在采样过程中的协助。
参考文献
Study on the admission levels of circulating cell-free DNA in patients with acute myocardial infarction using different quantification methods
DOI:10.1080/00365513.2019.1672086 URL [本文引用: 1]
A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide
We describe a modification of the DNA extraction method, in which cetyltrimethylammonium bromide (CTAB) is used to extract nucleic acids from plant tissues. In contrast to the original method, the modified CTAB procedure is faster, omits the selective precipitation and CsCl gradient steps, uses less expensive and toxic reagents, requires only inexpensive laboratory equipment and is more readily adapted to high-throughput DNA extraction. This protocol yields approximately 5-30 microg of total DNA from 200 mg of tissue fresh weight, depending on plant species and tissue source. It can be completed in as little as 5-6 h.
Genetic diversity and core collection for potato (Solanum tuberosum L.) cultivars from Cameroon as revealed by SSR markers
DOI:10.1007/s12230-017-9584-2 URL [本文引用: 1]
Construction of a genetic linkage map in man using restriction fragment length polymorphisms
We describe a new basis for the construction of a genetic linkage map of the human genome. The basic principle of the mapping scheme is to develop, by recombinant DNA techniques, random single-copy DNA probes capable of detecting DNA sequence polymorphisms, when hybridized to restriction digests of an individual's DNA. Each of these probes will define a locus. Loci can be expanded or contracted to include more or less polymorphism by further application of recombinant DNA technology. Suitably polymorphic loci can be tested for linkage relationships in human pedigrees by established methods; and loci can be arranged into linkage groups to form a true genetic map of "DNA marker loci." Pedigrees in which inherited traits are known to be segregating can then be analyzed, making possible the mapping of the gene(s) responsible for the trait with respect to the DNA marker loci, without requiring direct access to a specified gene's DNA. For inherited diseases mapped in this way, linked DNA marker loci can be used predictively for genetic counseling.
A framework for variation discovery and genotyping using next-generation DNA sequencing data
DOI:10.1038/ng.806
PMID:21478889
[本文引用: 1]

Recent advances in sequencing technology make it possible to comprehensively catalog genetic variation in population samples, creating a foundation for understanding human disease, ancestry and evolution. The amounts of raw data produced are prodigious, and many computational steps are required to translate this output into high-quality variant calls. We present a unified analytic framework to discover and genotype variation among multiple samples simultaneously that achieves sensitive and specific results across five sequencing technologies and three distinct, canonical experimental designs. Our process includes (i) initial read mapping; (ii) local realignment around indels; (iii) base quality score recalibration; (iv) SNP discovery and genotyping to find all potential variants; and (v) machine learning to separate true segregating variation from machine artifacts common to next-generation sequencing technologies. We here discuss the application of these tools, instantiated in the Genome Analysis Toolkit, to deep whole-genome, whole-exome capture and multi-sample low-pass (∼4×) 1000 Genomes Project datasets.
No reduction in genetic diversity of Swiss stone pine (Pinus cembra L.) in Tatra Mountains despite high fragmentation and small population size
DOI:10.1007/s10592-014-0628-6 URL [本文引用: 2]
STRUCTURE HARVESTER: A website and program for visualizing structure output and implementing the Evanno method
DOI:10.1007/s12686-011-9548-7 URL [本文引用: 1]
Growth patterns and genetic structure of Populus euphratica Oliv. (Salicaceae) forests in NW China-Implications for conservation and management
DOI:10.1016/j.foreco.2013.02.009 URL [本文引用: 2]
Phylogeography reveals geographic and environmental factors driving genetic differentiation of Populus sect. Turanga in northwest China
DOI:10.3389/fpls.2021.705083 URL [本文引用: 2]
Relationship between diameter at breast height and age of endangered species Populus euphratica Oliv
濒危物种胡杨胸径与树龄关系研究
Relationships between life history characteristics and electrophoretically detectable genetic variation in plants
DOI:10.1146/ecolsys.1979.10.issue-1 URL [本文引用: 1]
Genome resequencing reveals demographic history and genetic architecture of seed salinity tolerance in Populus euphratica
DOI:10.1093/jxb/eraa172 URL [本文引用: 5]
A comparative assessment of SNP and microsatellite markers for assigning parentage in a socially monogamous bird
DOI:10.1111/1755-0998.12589
PMID:27488248
[本文引用: 1]

Single-nucleotide polymorphisms (SNPs) are preferred over microsatellite markers in many evolutionary studies, but have only recently been applied to studies of parentage. Evaluations of SNPs and microsatellites for assigning parentage have mostly focused on special cases that require a relatively large number of heterozygous loci, such as species with low genetic diversity or with complex social structures. We developed 120 SNP markers from a transcriptome assembled using RNA-sequencing of a songbird with the most common avian mating system-social monogamy. We compared the effectiveness of 97 novel SNPs and six previously described microsatellites for assigning paternity in the black-throated blue warbler, Setophaga caerulescens. We show that the full panel of 97 SNPs (mean H = 0.19) was as powerful for assigning paternity as the panel of multiallelic microsatellites (mean H = 0.86). Paternity assignments using the two marker types were in agreement for 92% of the offspring. Filtering individual samples by a 50% call rate and SNPs by a 75% call rate maximized the number of offspring assigned with 95% confidence using SNPs. We also found that the 40 most heterozygous SNPs (mean H = 0.37) had similar power to assign paternity as the full panel of 97 SNPs. These findings demonstrate that a relatively small number of variable SNPs can be effective for parentage analyses in a socially monogamous species. We suggest that the development of SNP markers is advantageous for studies that require high-throughput genotyping or that plan to address a range of ecological and evolutionary questions.© 2016 John Wiley & Sons Ltd.
Genetic diversity of marginal populations of Populus euphratica Oliv. from highly fragmented river ecosystems
DOI:10.2478/sg-2020-0019 URL [本文引用: 1]
Conservation genetics of Pinus bungeana-Evaluation and conservation of natural populations’ genetic diversity
白皮松保育遗传学--天然群体遗传多样性评价与保护策略
Sampling strategies of sweet potato core collection based on morphological traits
基于形态性状的甘薯核心种质取样策略研究
Study on sampling schemes of core collection of local varieties of rice in Yunnan, China
云南地方稻种资源核心种质取样方案研究
Constructing a core collection of the medicinal plant Angelica biserrata using genetic and metabolic data
DOI:10.3389/fpls.2020.600249 URL [本文引用: 1]
Ancient polymorphisms and divergence hitchhiking contribute to genomic islands of divergence within a poplar species complex
A reciprocal transplant trial suggests a disadvantage of northward seed transfer in survival and growth of Japanese red pine (Pinus densiflora) trees
Genetic distance between populations
DOI:10.1086/282771 URL [本文引用: 1]
Analysis of gene diversity in subdivided populations
GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-An update
GenAlEx: Genetic Analysis in Excel is a cross-platform package for population genetic analyses that runs within Microsoft Excel. GenAlEx offers analysis of diploid codominant, haploid and binary genetic loci and DNA sequences. Both frequency-based (F-statistics, heterozygosity, HWE, population assignment, relatedness) and distance-based (AMOVA, PCoA, Mantel tests, multivariate spatial autocorrelation) analyses are provided. New features include calculation of new estimators of population structure: G'(ST), G''(ST), Jost's D(est) and F'(ST) through AMOVA, Shannon Information analysis, linkage disequilibrium analysis for biallelic data and novel heterogeneity tests for spatial autocorrelation analysis. Export to more than 30 other data formats is provided. Teaching tutorials and expanded step-by-step output options are included. The comprehensive guide has been fully revised.GenAlEx is written in VBA and provided as a Microsoft Excel Add-in (compatible with Excel 2003, 2007, 2010 on PC; Excel 2004, 2011 on Macintosh). GenAlEx, and supporting documentation and tutorials are freely available at: http://biology.anu.edu.au/GenAlEx.rod.peakall@anu.edu.au.
Conservation and restoration of pine forest genetic resources in Mexico
Genetic diversity of Populus euphratica populations in northwestern China determined by RAPD DNA analysis
DOI:10.1023/A:1015605928414 URL [本文引用: 1]
Molecular characterization of Nicaraguan Pinus tecunumanii Schw. ex Eguiluz et Perry populations for in situ conservation
DOI:10.1007/s00468-014-1005-2 URL [本文引用: 1]
Speciation of two desert poplar species triggered by Pleistocene climatic oscillations
DOI:10.1038/hdy.2013.87
PMID:24065180
[本文引用: 2]

Despite the evidence that the Pleistocene climatic fluctuations have seriously affected the distribution of intraspecific diversity, less is known on its impact on interspecific divergence. In this study, we aimed to test the hypothesis that the divergence of two desert poplar species Populus euphratica Oliv. and P. pruinosa Schrenk. occurred during the Pleistocene. We sequenced 11 nuclear loci in 60 individuals from the two species to estimate the divergence time between them and to test whether gene flow occurred after species separation. Divergence time between the two species was estimated to be 0.66-1.37 million years ago (Ma), a time at which glaciation was at its maximum in China and deserts developed widely in central Asia. Isolation-with-Migration model also indicated that the two species had diverged in the presence of gene flow. We also detected evidence of selection at GO in P. euphratica and to a lesser extent at PhyB2. Together, these results underscore the importance of Pleistocene climate oscillations in triggering plant speciation as a result of habitats divergence.
Genetic differentiation and delimitation between ecologically diverged Populus euphratica and P. pruinosa
DOI:10.1371/journal.pone.0026530 URL [本文引用: 3]
Strategy for the construction of a core collection for Pinus yunnanensis Franch. to optimize timber based on combined phenotype and molecular marker data
DOI:10.1007/s10722-021-01182-9 URL [本文引用: 2]
Development and characterization of microsatellite markers in Populus euphratica (Populaceae)
DOI:10.1111/men.2008.8.issue-5 URL [本文引用: 1]
Two highly validated SSR multiplexes (8-plex) for Euphrates’ poplar, Populus euphratica (Salicaceae)
DOI:10.1111/men.2012.13.issue-1 URL [本文引用: 1]
GCTA: A tool for genome-wide complex trait analysis
DOI:10.1016/j.ajhg.2010.11.011 URL [本文引用: 1]
A preliminary study on construction of high-quality core collection of Populus nigra
欧洲黑杨优质核心种质库的初步构建
Phylogeographic patterns of the desert poplar in Northwest China shaped by both geology and climatic oscillations
Genetic diversity of Populus pruinosa populations in Xinjiang of China based on SSR analysis
中国新疆灰叶胡杨群体遗传多样性的SSR分析
Analysis of drought characteristics in Xinjiang area based on remote sensing DSI index
基于遥感DSI的新疆干旱特征分析