



生物多样性 ›› 2009, Vol. 17 ›› Issue (3): 248-256. DOI: 10.3724/SP.J.1003.2009.08316 cstr: 32101.14.SP.J.1003.2009.08316
收稿日期:2008-12-04
接受日期:2009-02-18
出版日期:2009-05-20
发布日期:2009-05-20
通讯作者:
张传生
作者简介:*E-mail: cszhang1976@126.com基金资助:
Liying Geng1, Chuansheng Zhang2,3,*( ), Lixin Du3
), Lixin Du3
Received:2008-12-04
Accepted:2009-02-18
Online:2009-05-20
Published:2009-05-20
Contact:
Chuansheng Zhang
摘要:
为探讨鸡pre-microRNA SNP的多态性及其可能的功能意义, 对鸡471个pre-microRNA区域的SNP位点进行了鉴定和生物信息学分析。结果表明: pre-microRNA的SNP多态性显著地低于其侧翼区(P<0.01=, 其种子区SNP变异对pre-microRNA二级结构稳定性的影响高于其他各区; microRNA成熟体SNP可能影响microRNA对靶基因的选择。研究结果提示: pre-microRNA相对于其侧翼区在分子进化过程中受到更大的选择压力; 成熟体SNP可通过影响microRNA加工和靶基因的选择, 改变多种生理过程, 导致鸡种间表型变异。研究结果将为鸡microRNA的进化模式研究及其功能性SNP的鉴定提供参考信息。
耿立英, 张传生, 杜立新 (2009) 鸡基因组pre-microRNA SNP多态性. 生物多样性, 17, 248-256. DOI: 10.3724/SP.J.1003.2009.08316.
Liying Geng, Chuansheng Zhang, Lixin Du (2009) Single nucleotide polymorphisms in chicken genomic pre-microRNA. Biodiversity Science, 17, 248-256. DOI: 10.3724/SP.J.1003.2009.08316.
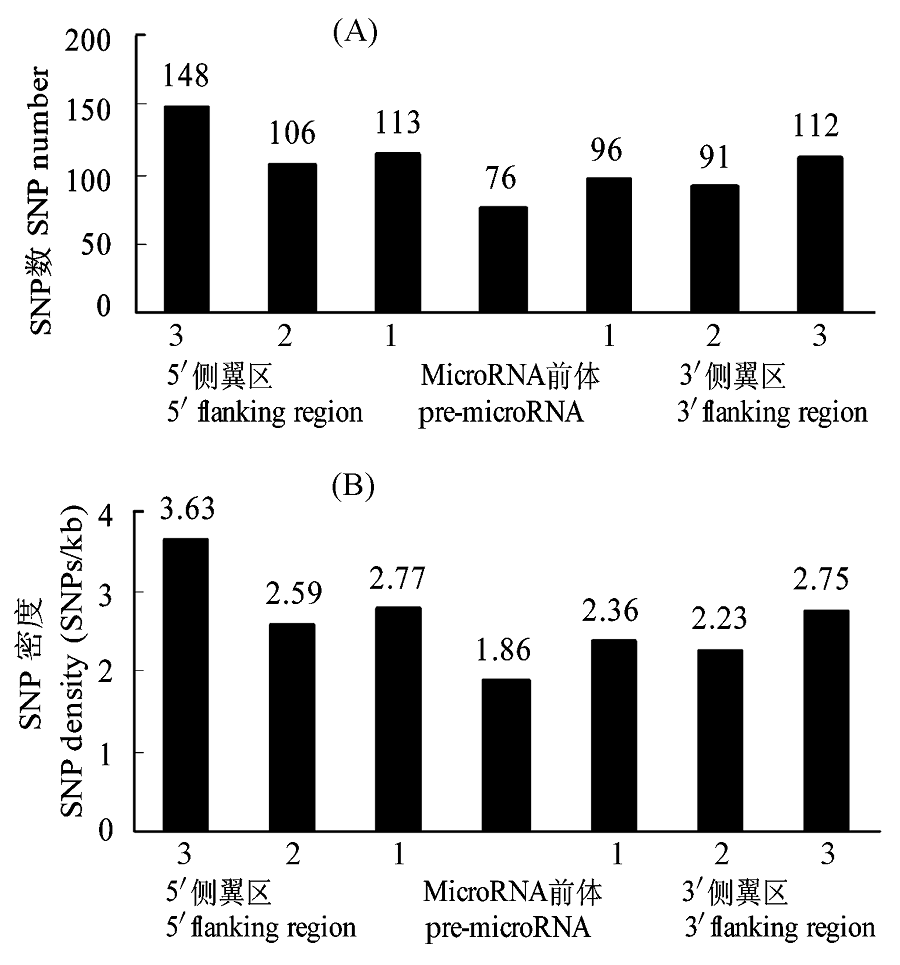
图2 鸡pre-microRNAs及其侧翼区的SNP的数量(A)及密度(B)。侧翼区1-3是指与pre-microRNAs邻接、无重叠的3个等长侧翼区。
Fig. 2 SNP in chicken pre-microRNAs. (A) SNP numbers in validated chicken pre-microRNAs and flanking regions. (B) SNP density in validated chicken pre-microRNAs and flanking regions. Flanking regions 1-3 represent successive, non- overlapping windows (equal to the size of the given pre-microRNA) located immediately adjacent to the pre-microRNA.
| 定位 Location | 基因 microRNA | SNP 登录号 dbSNP | 变异 Variation | Pre-microRNA序列变异 Variation of the pre-microRNA sequence | 自由能改变值 |
|---|---|---|---|---|---|
| ΔΔG kcal/mol | |||||
| Loop | gga-mir-1558 | rs14268121 | GCCCT/- | CGCTGCTCCAGAGCCCGTTATACTCAGCAGGACAAGAGGGGAC[AGGC/-]A GGGCTGCTCACTGCTGTGATGGGAGCTCTGGGATGCG | 1.9 |
| Loop | gga-mir-133b | rs16341692 | A/G | CTCTGCTCTGGCTGGTCAAACGGAACCAAGCCCGTCTTC[T/C]TCGGAGGTT TGGTCCCCTTCAACCAGCTATAGCAGTGTTGAAAA | 1.4 |
| Loop | gga-mir-124a | rs14237202 | -/A | AGGCTCTGCCTCTCCGTGTTCACAGCGGACCTTGATTTAA[A/-]TCATACAA TTAAGGCACGCGGTGAATGCCAAGAGCGGATCCTCCAGGCGGCATT | 0.1 |
| Loop | gga-mir-1560 | rs14968200 | G/A | TCCTTAGTCACTGCGTGGGGGTGGCGGCGCGAGCAGAGAGGCGCTGCC[G/A ]TGTCCAAAGCAGCATCTCTGGACGCGCTCGTTCCTTCTGCGTGGTCCGCTT GGA | 0 |
| Loop | gga-mir-1660 | rs16284368 | T/C | CAAGTAAACATAAAGGCATAGTATAC[G/A]ATAGGCC TGTGTTTTTGTGCA TATTTG | 0 |
| Loop | gga-mir-1816 | rs16057773 | A/G | GGATGGTGAAATGATCTGAAGCTTGTAGGTTTTGTGGTTTTGTTTGACGAT T[A/G]CATGTTAAATTTAATCATCTCAACATGCATCCATCAGTGTTTTACTAATCT | 0 |
| MIRΔseed | gga-mir-1550- 5p | rs14981477 | -/G | CCGACCTGTGCAGGCTTGGGCTGATGGGGG[G/-]TGTAGTGCAGCAGCTATTTATCATGCTTCACTTACACTGCTCACTGGTCTAAGCTGCACCACCCGGG | 4.0 |
| MIRΔseed | gga-mir-1712* | rs15403608 | T/C | GCCCTTCTGGCTCCAGTCTTAACT[A/G]CAGGCAATGCTGACATTCTCCTTCAGTTATCAGTGGAGTTTGGAGTGC | 3.8 |
| MIRΔseed | gga-mir-1675 | rs16643637 | T/C | GCCTTCCCGCTTGGCCCCGCTCCCGCTCAGGGCGGTGTCAGGCGGCGCCGCCGGACGCCATCTTG[A/G]CTGTGGGCAGCTCGGCTTCAGGTCGGCAGAGAGC | 2.9 |
| MIRΔseed | gga-mir-455-5p | rs15031616 | -/G | TCCCTGGTGTGAGGGTATGTGCCC[C/-]TTGGACTACATCGTGGAAGCCAGCACCATGCAGTCCATGGGCATATACACTTGCCTCAAGGT | 1.0 |
| MIRΔseed | gga-mir-1648* | rs14281066 | G/A | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGCGTGGTGGGAGTGGA[G/A]CGGAGCTGTCCGCCGTAGCCG | 0.6 |
| MIRΔseed | gga-mir-1596* | rs15190357 | C/T | TGGACTTTGTTTCATTCTCCTTCAATCCATCTTTGTCATGATTTACAAAAAG AACAAAGGTGGCC[G/A]TGGGTGAAT GAAGCCAGGCATCCA | 0.5 |
| MIRΔseed | gga-mir-1568 | rs14511526 | T/C | GCAACTGCCTTGTGATGGGTGAGTTTTCACTTTTTTTTCCTGGACTCATAG[A /G]TCTGAAGGCAGGTAGC | 0.3 |
| MIRΔseed | gga-mir-1703 | rs16718265 | C/- | AGCTTGCCGCAGGATTCAGAGGCTGTA[G/-]GTCCCGTGCTTTAATTAAAAG AACGAAGCTCGTGACTTCAGCTCCAGCATCCGTGGCAGCT | 0.3 |
| Seed | gga-mir-1644 | rs14076349 | T/C | GTGTCCCCAGGTAGCGCTGAGCCCAGCAGGCAGTGCTGTGCCAATCTGGC TGCTC[T/C]GTTGTGCAGGGCTGTGCTCCCAGGGTGTGC | 7.0 |
| Seed | gga-mir-1658* | rs16681031 | C/G | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGT TTCTCAGAGCT[G/C]TGGGTTGGTGTTGATGGGGAGCTGGGG | 6.2 |
| Seed | gga-mir-1648* | rs14281065 | T/- | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGC GTGGTGGGAG[T/-]GGAGCGGAGCTGTCCGCCGTAGCCG | 4.4 |
| Seed | gga-mir-1658 | rs16681033 | -/G | CCCCCTTCCTGAATATACCA[G/-]CCCCCAGGAGTTCTGCCATACCAGTGTG TGTTTCTCAGAGCTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 4.2 |
| Seed | gga-mir-1614 | rs15172520 | G/A | GCTGGAGTTGGGTGGCATGGCAGACTCACCCTGCAGCTCTGACACAGGGC AGG[G/A]AGGAACTGCCAGCAGACTCAGCTCCAGC | 4.0 |
| Seed | gga-mir-1568 | rs14511527 | A/G | GCAACTGCCTTGTGATGGGTGAGTTTTCACTTTTTTTTCCTGGACTCA[C/T]A GATCTGAAGGCAGGTAGC | 1.5 |
| Seed | gga-mir-1657 | rs14934924 | G/A | GAGAAGTATATGTGTATACATCAACACATACTTATATTTGTATATGGATTTA CATATAT[G/A]TGTATTGTTGGTGTGTGA TACGATATTCCATATC | 0.3 |
| Seed | gga-mir-1658* | rs16681032 | C/T | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGTT TCTCAGA[G/A]CTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 0 |
| MIR* | gga-mir-1658 | rs16681031 | C/G | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGTT TCTCAGAGCT[G/C]TGGGTTGGTGTTGATGGGGAGCTGGGG | 6.2 |
| MIR* | gga-mir-1652 | rs16744833 | C/G | GGGGAAGGCCCGTGGGGATGCTCCTACCCGGG[G/C]CGCGGGGCTGCTGTCCCCACCATCCCTGTGTCCCCTCGGTGGCAGCAGGACTCTGCCATCCCC | 5.4 |
| MIR* | gga-mir-1648 | rs14281065 | T/- | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGC GTGGTGGGAG[T/-]GGAGCGGAGCTGTCCGCCGTAGCCG | 4.4 |
| MIR* | gga-mir-1658* | rs16681033 | -/G | CCCCCTTCCTGAATTACCA[G/-]CCCCCAGG AGTTCTGCCATACCAGTGTGT GTTTCTCAGAGCTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 4.2 |
| 定位 Location | 基因 microRNA | SNP 登录号 dbSNP | 变异 Variation | Pre-microRNA序列变异 Variation of the pre-microRNA sequence | 自由能改变值 |
| ΔΔG (kcal/mol) | |||||
| MIR* | gga-mir-1550-3p | rs14981477 | -/G | CCAGACCTGTGCAGGCTTGGGCTGATGGGGG[G/-]TGTAGTGCAGCAGCTAT TTATCATGCTTCACTTACACTGCTCACTGGTCTAAGCTGCACCACCCGGG | 4.0 |
| MIR* | gga-mir-1614 | rs15172520 | G/A | GCTGGAGTTGGGTGGCATGGCAGACTCACCCTGCAGCTCTGACACAGGGC AGG[G/A]AGGAACTGCCAGCAGACTCAGCTCCAGC | 4.0 |
| MIR* | gga-mir-1656 | rs13942534 | -/T | TCTCTGTCACCAGCGGGCATTGGCATCACTGGCATGGCTGTG[T/-]ATGTCA TGAGTGCTGGCAGCAGGA | 3.8 |
| MIR* | gga-mir-1712 | rs15403608 | T/C | GCCCTTCTGGCTCCAGTCTTAACT[A/G]CAGGCAATGCTGACATTCTCCTTCAGTTATCAGTGGAGTTTGGAGTGC | 3.8 |
| MIR* | gga-mir-449 | rs14734667 | G/A | CTGTGTGCGGTGGGGTGGCAGTGTATGTTAGCTGGTTGAAACTCTTGACAT CAGCTAACA[C/T]GCAGTTGCTAACCTGCTCCACATAC | 3.3 |
| MIR* | gga-mir-1659 | rs15847005 | G/A | CACTAGTTTCATATCCAGAGGAAGACTTCATTTTATTTAATGTT[C/T]TCATT TCACTGAATAGGGCAGAATGAAGGAGTCCCTGCCTGTGGTGAGTGACAGTG | 2.6 |
| MIR* | gga-mir-1683 | rs15262856 | A/C | GCTGTGCCAGGCTCCACTGGCGATGCTT[T/G]TTGTCACCTGGATGTGAGATGGTCAGTTCTGGGACAGTCCAGCATCTTTCTGGTTTATGCTGTGTCAGC | 2.4 |
| MIR* | gga-mir-1610 | rs16628188 | T/C | CATGGCGCCGCTCG[A/G]GCCCGCAGAGGCCGCCGGGAGCTGCAGTGCGCGCGGCATGGCTTGTGGTGGAACGGGCGGCTCCCCTGTCGTG | 1.6 |
| MIR* | gga-mir-1783 | rs14039125 | CT/- | CTGAAGTGTTATGAAATCGTGCTCTGAACGAGCTGAGAGGCATGCTACTTTCTGATCATTGCATCCAG[CT/-]CTCTCATTGCTCATTTCATAACTCCTTTCAG | 1.4 |
| MIR* | gga-mir-138-1 | rs15960797 | C/A | CCCTGCCGGGTGCCGTGCAGCAGCTGGTGTTGTGAATCAGGCCGTCACCAGTCGGAGAACGGCTACTTCACAACACCAG[G/T]GTGGCACTGCACCACA | 1.3 |
| MIR* | gga-mir-1605 | rs15600725 | G/A | AAGAGAAGTTCACAGAGGGTTTCATGTGTATCTCTTGAAGATGCTGAGTATGAGAGAAGCATATGAAAG[C/T]GCTTCTGTGATTTTTCTT | 1.3 |
| MIR* | gga-mir-455-3p | rs15031616 | -/G | TCCCTGGTGTGAGGGTATGTGCCC[C/-]TTGGACTACATCGTGGAAGCCAGCACCATGCAGTCCATGGGCATATACACTTGCCTCAAGGT | 1.0 |
| MIR* | gga-mir-1783 | rs14039126 | A/G | CTGAAGTGTTATGAAATCGTGCTCTGAACGAGCTGAGAGGCATGCTACTTTCTGATCATTGCATCAGCTCTCTC[A/G]TTGCTCATTTCATAACTCCTTTCAG | 0.7 |
| MIR* | gga-mir-16-1 | rs15498115 | T/- | GTCTGTCATACTCTAGCAGCACGTAAATATTGGTGTTAAAACTGTAAATATC TCCAGTATTA[A/-]CTGTGCTGCTGAAGTAAGGCT | 0.6 |
| MIR* | gga-mir-1648 | rs14281066 | G/A | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGCGTGGTGGGAGTGGA[G/A]CGGAGCTGTCCGCCGTAGCCG | 0.6 |
| MIR* | gga-mir-1719 | rs14969646 | C/G | TTGAGAAGTACTTGTTTGTACCAAAAGATGATGAGTTGCTGATGTGCTTTGTCGAGGTAACGCT[G/C]TTTCCTGGGTGCAGTATCGCTTAA | 0.6 |
| MIR* | gga-mir-1596 | rs15190357 | C/T | TGGACTTTGTTTCATTCTCCTTCAATCCATCTTTGTCATGATTTACAAAAAG AACAAAGGTGGCC[G/A]TGGGTGAATGAAGCCAGGCATCCA | 0.5 |
| MIR* | gga-mir-1558 | rs14268122 | A/G | CGCTGCTCCGAGCCCGTTATA[T/C]TCAGCAGGACAAGAGGGGACAGGCAGGGCTGCTCACTGCTGTGATGGGAGCTCTGGGATGCG | 0.2 |
| MIR* | gga-mir-1605 | rs15600727 | T/C | AAGAGAAGTTCACAGAGGGTTTCATGTGTATCTCTTGAAGATGCTGATATG AGAGA[A/G]GCATATGAAAGCGCTTCTGTGATTTTTCTT | 0 |
| MIR* | gga-mir-1606 | rs14431112 | C/G | TGTGAGGCTGCTCTT[G/C]GGTTTCCCTCTGAAATTCACTGAAAGAGGGGGCAACAGAACAGTCCTGAACA | 0 |
| MIR* | gga-mir-1618 | rs14639002 | C/G | CTGCGTGCTGCATCCTGGCTCCTGG[C/G]GTCCTGCACGTGCTATTGGAGAGCCCGGAGCCAGGCTGCTGTTCTCCAGGCAG | 0 |
| MIR* | gga-mir-1630 | rs16651110 | T/C | TTTGGGCTTT[C/T]GTTTCTTCTTTCTAAAGGGGGAGGAAAGAGGAAACTGAAGCTGAAA | 0 |
| MIR* | gga-mir-1658 | rs16681032 | C/T | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGTTTCTCAGA[G/A]CTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 0 |
| Other | gga-mir-1600 | rs14726712 | T/A | GCCCTTCTGGCTCCAGTCTTAACT[A/G]CAGGCAATGCTGACATTCTCCTTCAGTTATCAGTGGAGTTTGGAGTGC | 4.6 |
| Other | gga-mir-1768 | rs13538425 | T/G | TATCAGCTCATCAGCATGACAATCCTCTGCTGCCTTGGGAGTTGTGAGCAGAGGATTGTCGTGCTGA[T/G]GAGTTGATA | 4.3 |
| Other | gga-mir-1562 | rs14653234 | A/T | ATCAACAAGCGGGGTGCAGCTGCACTGTAGATGGAGGGGCTGCATGAAGGTGGATAGTTTCTGTGCCCCGTGTATGCAGTGCAGGTGCTC[T/T]CTGCACTT GGT | 4.1 |
| Other | gga-mir-1453 | rs16159272 | G/A | GTGCAGGCTCTCTGTGGCTCAGCCCAGCTG[T/C]GGTACTGCATGGTGCTGGGCAGCCTCTCAGTGAGCTCTGTGCT | 3.8 |
| 定位 Location | 基因 microRNA | SNP 登录号 dbSNP | 变异 Variation | Pre-microRNA序列变异 Variation of the pre-microRNA sequence | 自由能改变值 |
| ΔΔG (kcal/mol) | |||||
| Other | gga-mir-1666 | rs14120863 | C/G | GCCAGGAGCAAACACGA[G/C]TTAACGCCACGGGGCTGAGGCTGAGCTGCA AACTTTCACAGCTTCAGAGCGTGGCTGCAGCGCGTGCTGGC | 3.5 |
| Other | gga-mir-1704 | rs14668705 | C/G | GACCGA[C/G]AATTCACAGCAGATTGATGGGGCACACCAAGATCTGTGCCT GTTCCTGCTGGGATGCTCCCGTC | 3.4 |
| Other | gga-mir-1792 | rs14317161 | A/T | TCCAAAGCCACCTCTATGCTGCGATAGCTCAGGAACACTGCAGTCAAGAACACATACTGGAGTGTTCCTGGGGTATCACAGCGTAGGGGTGGT[T/A]TTGGA | 3.2 |
| Other | gga-mir-1812 | rs16571431 | A/G | CTTTATTGGGAACCTAGATATATACTGTATACTGTGTCAAGGTTTAGATGACATACTATACAGTATATATCTAGGTT[T/C]CTAATACAAG | 2.9 |
| Other | gga-mir-1608 | rs16662849 | A/G | TCTCATCTGGTGCCCCCGGGATGGGACAGTGGCTGCGCCCTCTGTGAAGGGAA[C/T]GGAGGGCTCAGCAGCTGTGCCCGTGCCTGGCAGAGTGGA | 2.5 |
| Other | gga-mir-1705 | rs15027520 | A/G | GCTGAGAGCTTTCCTCAACTGTGTGCTGATTTCATTAGACTTTTTCCTGACAAGAAATCTGGAAGTCAGCACATGCTGAGAAGA[G/A]TCTGC | 2.1 |
| Other | gga-mir-1757 | rs15505339 | G/C | GCTGTTTTGCCGTGGATGAACAAAACTCTTCTGGTCCGCTGTTTAGTTCAATTAGCGCTTCAGAGGCGGAACAGTAGGTTTCTGTTCTT[C/G]CAGCCAAGATTCTGT | 1.8 |
| Other | gga-mir-1749 | rs15860000 | C/T | GAGTGCAGCTGGAGGGCTACAGGGAGATAAGGAACTGAACTGGCTGCATCTCTGCTGCTTGGCTCTGTTCCCTATTTCC[C/T]GTCTCCTTAGCAAATCTC | 1.5 |
| Other | gga-mir-1657 | rs14934923 | A/G | GAGAAGTAT[A/G]TGTGTATACATCAACACATACTTATATTTGTATATGGAT TTACATATATGTGTATTGTTGGTGTGTGATACGATATTCCATATC | 1.2 |
| Other | gga-mir-1562 | rs14653232 | A/G | ACAACA[A/G]GCGGGGTGCAGCTGCACTGTAGATGGAGGGGCTGCATGAAGGTGGATAGTTTCTGTGCCCCGTGTATGCAGTGCAGGTGCTCACTGCACTGGT | 1.1 |
| Other | gga-mir-1704 | rs14668704 | C/T | GACCGAGAATTCACAGCAGATTGATGGGGCACACCAAGATCTGTGCCTGTTCCTGCTGGGATGCTCCC[G/A]TC | 1.0 |
| Other | gga-mir-1558 | rs15169308 | G/A | CGCTGCTCCAGAGCCCGTTATACTCAGCAGGACAAGAGGGGACAGGCAGGGCTGCTCACTGCTGTGATGGGAGCTCTGGGATG[C/T]G | 0.9 |
| Other | gga-mir-1790 | rs16361960 | G/A | GGCGGGGCGGGATGCCGCTGGGGC[T/C]GGGCCTCGTGCCGTGCTCTGCTGGGCCGGAGAAGGCGGGTGAGGGCGGTGGGGGAAGGGAAGCGGAGCTGTTTCTTCGCT | 0.8 |
| Other | gga-mir-1675 | rs16643638 | C/T | GCCTTCCC[G/A]CTTGGCCCCGCTCCCGCTCAGGGCGGTGTCAGGCGGCGCCGCCGGACGCCATCTTGACTGTGGGCAGCTCGGCTTCAGGTCGGCAGAGAGC | 0.7 |
| Other | gga-mir-551 | rs16677358 | A/G | CCACCAGAGC[T/G]GCCCCATGGCTCCAGAAATCAAGGGTGGGTAAGACCT TGTAGGATAACTGGCAGGCGACCCATACTTGGTTTCAGGGGCTGTGTGGGTATCG- TCAGTGGTGG | 0.6 |
| Other | gga-mir-1762 | rs13608151 | T/C | GGTGCTCCTCCAGCTGCAGGGCAGGAGGGAAGCAGAGCTTAGAGCTGTTTTCTACGCTCGAGCTGGCTTTGCTCAAGCCCCGTGTGCGG[A/G]GGGTTTCC | 0.5 |
| Other | gga-mir-1608 | rs15961488 | A/C | TCTCATCTGGTGCCCCCGGGATGGGACAGTGGCTGCGCCCTCTGTGAAGGGAATGGAGGGCTCAGCAGCTGTGCCCGTGCCTGGCAGAG[T/G]GGA | 0.5 |
| Other | gga-mir-1719 | rs14969645 | A/G | TTGAGAAGTACTTGTTTGTACCAAAAGATGATGAGTTGCTGATGTGCTTTGTCGAGGTAACGCTGTTTCCTGGG[T/C]GCAGTATCGCTTA | 0.4 |
| Other | gga-mir-1458 | rs16654946 | A/- | TTAGTTCAGCCTCA[T/-]TTTTTTTTAGAGTTCTGGAGTATCTTATTGCACAAAATATGCTGAGGAGAAATTCCTGTGATGCTCATGAGAAAAATTGTAGAGCGCTTTTACACT | 0.4 |
| Other | gga-mir-1562 | rs14653233 | G/A | ACAACAAGCGGG[G/A]TGCAGCTGCACTGTAGATGGAGGGGCTGCATGAAGGTGGATAGTTTCTGTGCCCCGTGTATGCAGTGCAGGTGCTCACTGCACTTGG GT | 0.4 |
| Other | gga-mir-1619 | rs14280616 | G/T | TTGTAAGTCCAGCAGTCTGTATTGCTACAAATGCTCTTCTCAGCGCAA[C/A] CTCAGAAGAATTCATAGCAATACGGACTGCTGGACTTCTAG | 0.3 |
| Other | gga-mir-1675 | rs16643636 | A/T | GCCTTCCCGCTTGGCCCCGCTCCCGCTCAGGGCGGTGTCAGGCGGCGCCG- CCGGACGCCATCTTGACTGTGGGCAGC[T/A]CGGCTTCAGGTCGGCAGAG- AGC | 0.3 |
| Other | gga-mir-1687 | rs15179830 | T/G | GATTTCTTTGTGTCTGTGGTCTGAGGAAATGAGCCAGCTGAGTTAGCAACTTCTTTGCTGGCTGTTTCTCAGTGACTGCAGCATAAAAA[T/G]C | 0.2 |
| Other | gga-mir-22 | rs14120771 | C/T | GCTCCTGGCTGCCC[G/A]GCAGCAGTTCTTCAGTGGCAAGCTTTATGTCCTTCTCTAGTAGCTAAAGCTGCCAGTTGAAGAACTGTTGAATGTAGCCACGTTC | 0 |
| Other | gga-mir-1659 | rs15847007 | G/A | CATAGTTTCATAT[C/T]CAGAGGAAGACTTCATTTTATTTAATGTTCTCATTTCACTGAATAGGGCAGAATGAAGGAG TCC CTGCCTGTGGTGAGTGACAGTG | 0 |
| Other | gga-mir-1590 | rs15781755 | A/G | TGGA[T/C]GGAAATAGGTATCTCCAGAGAACAGTTTGTCCCACTTGTTTTTGACGTCTGTCGTAGCAAGGGACAAATTGCTCTCTGGAAGTACATTTCTCCA | 0 |
表1 SNP对pre-microRNA二级结构的影响(方括号内显示的为pre-microRNA变异位点, 下划线标出的为microRNA成熟体)
Table 1 Effect of the SNP on the secondary structure of pre-microRNA. The polymorphic sites are showed in brackets, and the mature of microRNA are underlined.
| 定位 Location | 基因 microRNA | SNP 登录号 dbSNP | 变异 Variation | Pre-microRNA序列变异 Variation of the pre-microRNA sequence | 自由能改变值 |
|---|---|---|---|---|---|
| ΔΔG kcal/mol | |||||
| Loop | gga-mir-1558 | rs14268121 | GCCCT/- | CGCTGCTCCAGAGCCCGTTATACTCAGCAGGACAAGAGGGGAC[AGGC/-]A GGGCTGCTCACTGCTGTGATGGGAGCTCTGGGATGCG | 1.9 |
| Loop | gga-mir-133b | rs16341692 | A/G | CTCTGCTCTGGCTGGTCAAACGGAACCAAGCCCGTCTTC[T/C]TCGGAGGTT TGGTCCCCTTCAACCAGCTATAGCAGTGTTGAAAA | 1.4 |
| Loop | gga-mir-124a | rs14237202 | -/A | AGGCTCTGCCTCTCCGTGTTCACAGCGGACCTTGATTTAA[A/-]TCATACAA TTAAGGCACGCGGTGAATGCCAAGAGCGGATCCTCCAGGCGGCATT | 0.1 |
| Loop | gga-mir-1560 | rs14968200 | G/A | TCCTTAGTCACTGCGTGGGGGTGGCGGCGCGAGCAGAGAGGCGCTGCC[G/A ]TGTCCAAAGCAGCATCTCTGGACGCGCTCGTTCCTTCTGCGTGGTCCGCTT GGA | 0 |
| Loop | gga-mir-1660 | rs16284368 | T/C | CAAGTAAACATAAAGGCATAGTATAC[G/A]ATAGGCC TGTGTTTTTGTGCA TATTTG | 0 |
| Loop | gga-mir-1816 | rs16057773 | A/G | GGATGGTGAAATGATCTGAAGCTTGTAGGTTTTGTGGTTTTGTTTGACGAT T[A/G]CATGTTAAATTTAATCATCTCAACATGCATCCATCAGTGTTTTACTAATCT | 0 |
| MIRΔseed | gga-mir-1550- 5p | rs14981477 | -/G | CCGACCTGTGCAGGCTTGGGCTGATGGGGG[G/-]TGTAGTGCAGCAGCTATTTATCATGCTTCACTTACACTGCTCACTGGTCTAAGCTGCACCACCCGGG | 4.0 |
| MIRΔseed | gga-mir-1712* | rs15403608 | T/C | GCCCTTCTGGCTCCAGTCTTAACT[A/G]CAGGCAATGCTGACATTCTCCTTCAGTTATCAGTGGAGTTTGGAGTGC | 3.8 |
| MIRΔseed | gga-mir-1675 | rs16643637 | T/C | GCCTTCCCGCTTGGCCCCGCTCCCGCTCAGGGCGGTGTCAGGCGGCGCCGCCGGACGCCATCTTG[A/G]CTGTGGGCAGCTCGGCTTCAGGTCGGCAGAGAGC | 2.9 |
| MIRΔseed | gga-mir-455-5p | rs15031616 | -/G | TCCCTGGTGTGAGGGTATGTGCCC[C/-]TTGGACTACATCGTGGAAGCCAGCACCATGCAGTCCATGGGCATATACACTTGCCTCAAGGT | 1.0 |
| MIRΔseed | gga-mir-1648* | rs14281066 | G/A | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGCGTGGTGGGAGTGGA[G/A]CGGAGCTGTCCGCCGTAGCCG | 0.6 |
| MIRΔseed | gga-mir-1596* | rs15190357 | C/T | TGGACTTTGTTTCATTCTCCTTCAATCCATCTTTGTCATGATTTACAAAAAG AACAAAGGTGGCC[G/A]TGGGTGAAT GAAGCCAGGCATCCA | 0.5 |
| MIRΔseed | gga-mir-1568 | rs14511526 | T/C | GCAACTGCCTTGTGATGGGTGAGTTTTCACTTTTTTTTCCTGGACTCATAG[A /G]TCTGAAGGCAGGTAGC | 0.3 |
| MIRΔseed | gga-mir-1703 | rs16718265 | C/- | AGCTTGCCGCAGGATTCAGAGGCTGTA[G/-]GTCCCGTGCTTTAATTAAAAG AACGAAGCTCGTGACTTCAGCTCCAGCATCCGTGGCAGCT | 0.3 |
| Seed | gga-mir-1644 | rs14076349 | T/C | GTGTCCCCAGGTAGCGCTGAGCCCAGCAGGCAGTGCTGTGCCAATCTGGC TGCTC[T/C]GTTGTGCAGGGCTGTGCTCCCAGGGTGTGC | 7.0 |
| Seed | gga-mir-1658* | rs16681031 | C/G | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGT TTCTCAGAGCT[G/C]TGGGTTGGTGTTGATGGGGAGCTGGGG | 6.2 |
| Seed | gga-mir-1648* | rs14281065 | T/- | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGC GTGGTGGGAG[T/-]GGAGCGGAGCTGTCCGCCGTAGCCG | 4.4 |
| Seed | gga-mir-1658 | rs16681033 | -/G | CCCCCTTCCTGAATATACCA[G/-]CCCCCAGGAGTTCTGCCATACCAGTGTG TGTTTCTCAGAGCTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 4.2 |
| Seed | gga-mir-1614 | rs15172520 | G/A | GCTGGAGTTGGGTGGCATGGCAGACTCACCCTGCAGCTCTGACACAGGGC AGG[G/A]AGGAACTGCCAGCAGACTCAGCTCCAGC | 4.0 |
| Seed | gga-mir-1568 | rs14511527 | A/G | GCAACTGCCTTGTGATGGGTGAGTTTTCACTTTTTTTTCCTGGACTCA[C/T]A GATCTGAAGGCAGGTAGC | 1.5 |
| Seed | gga-mir-1657 | rs14934924 | G/A | GAGAAGTATATGTGTATACATCAACACATACTTATATTTGTATATGGATTTA CATATAT[G/A]TGTATTGTTGGTGTGTGA TACGATATTCCATATC | 0.3 |
| Seed | gga-mir-1658* | rs16681032 | C/T | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGTT TCTCAGA[G/A]CTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 0 |
| MIR* | gga-mir-1658 | rs16681031 | C/G | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGTT TCTCAGAGCT[G/C]TGGGTTGGTGTTGATGGGGAGCTGGGG | 6.2 |
| MIR* | gga-mir-1652 | rs16744833 | C/G | GGGGAAGGCCCGTGGGGATGCTCCTACCCGGG[G/C]CGCGGGGCTGCTGTCCCCACCATCCCTGTGTCCCCTCGGTGGCAGCAGGACTCTGCCATCCCC | 5.4 |
| MIR* | gga-mir-1648 | rs14281065 | T/- | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGC GTGGTGGGAG[T/-]GGAGCGGAGCTGTCCGCCGTAGCCG | 4.4 |
| MIR* | gga-mir-1658* | rs16681033 | -/G | CCCCCTTCCTGAATTACCA[G/-]CCCCCAGG AGTTCTGCCATACCAGTGTGT GTTTCTCAGAGCTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 4.2 |
| 定位 Location | 基因 microRNA | SNP 登录号 dbSNP | 变异 Variation | Pre-microRNA序列变异 Variation of the pre-microRNA sequence | 自由能改变值 |
| ΔΔG (kcal/mol) | |||||
| MIR* | gga-mir-1550-3p | rs14981477 | -/G | CCAGACCTGTGCAGGCTTGGGCTGATGGGGG[G/-]TGTAGTGCAGCAGCTAT TTATCATGCTTCACTTACACTGCTCACTGGTCTAAGCTGCACCACCCGGG | 4.0 |
| MIR* | gga-mir-1614 | rs15172520 | G/A | GCTGGAGTTGGGTGGCATGGCAGACTCACCCTGCAGCTCTGACACAGGGC AGG[G/A]AGGAACTGCCAGCAGACTCAGCTCCAGC | 4.0 |
| MIR* | gga-mir-1656 | rs13942534 | -/T | TCTCTGTCACCAGCGGGCATTGGCATCACTGGCATGGCTGTG[T/-]ATGTCA TGAGTGCTGGCAGCAGGA | 3.8 |
| MIR* | gga-mir-1712 | rs15403608 | T/C | GCCCTTCTGGCTCCAGTCTTAACT[A/G]CAGGCAATGCTGACATTCTCCTTCAGTTATCAGTGGAGTTTGGAGTGC | 3.8 |
| MIR* | gga-mir-449 | rs14734667 | G/A | CTGTGTGCGGTGGGGTGGCAGTGTATGTTAGCTGGTTGAAACTCTTGACAT CAGCTAACA[C/T]GCAGTTGCTAACCTGCTCCACATAC | 3.3 |
| MIR* | gga-mir-1659 | rs15847005 | G/A | CACTAGTTTCATATCCAGAGGAAGACTTCATTTTATTTAATGTT[C/T]TCATT TCACTGAATAGGGCAGAATGAAGGAGTCCCTGCCTGTGGTGAGTGACAGTG | 2.6 |
| MIR* | gga-mir-1683 | rs15262856 | A/C | GCTGTGCCAGGCTCCACTGGCGATGCTT[T/G]TTGTCACCTGGATGTGAGATGGTCAGTTCTGGGACAGTCCAGCATCTTTCTGGTTTATGCTGTGTCAGC | 2.4 |
| MIR* | gga-mir-1610 | rs16628188 | T/C | CATGGCGCCGCTCG[A/G]GCCCGCAGAGGCCGCCGGGAGCTGCAGTGCGCGCGGCATGGCTTGTGGTGGAACGGGCGGCTCCCCTGTCGTG | 1.6 |
| MIR* | gga-mir-1783 | rs14039125 | CT/- | CTGAAGTGTTATGAAATCGTGCTCTGAACGAGCTGAGAGGCATGCTACTTTCTGATCATTGCATCCAG[CT/-]CTCTCATTGCTCATTTCATAACTCCTTTCAG | 1.4 |
| MIR* | gga-mir-138-1 | rs15960797 | C/A | CCCTGCCGGGTGCCGTGCAGCAGCTGGTGTTGTGAATCAGGCCGTCACCAGTCGGAGAACGGCTACTTCACAACACCAG[G/T]GTGGCACTGCACCACA | 1.3 |
| MIR* | gga-mir-1605 | rs15600725 | G/A | AAGAGAAGTTCACAGAGGGTTTCATGTGTATCTCTTGAAGATGCTGAGTATGAGAGAAGCATATGAAAG[C/T]GCTTCTGTGATTTTTCTT | 1.3 |
| MIR* | gga-mir-455-3p | rs15031616 | -/G | TCCCTGGTGTGAGGGTATGTGCCC[C/-]TTGGACTACATCGTGGAAGCCAGCACCATGCAGTCCATGGGCATATACACTTGCCTCAAGGT | 1.0 |
| MIR* | gga-mir-1783 | rs14039126 | A/G | CTGAAGTGTTATGAAATCGTGCTCTGAACGAGCTGAGAGGCATGCTACTTTCTGATCATTGCATCAGCTCTCTC[A/G]TTGCTCATTTCATAACTCCTTTCAG | 0.7 |
| MIR* | gga-mir-16-1 | rs15498115 | T/- | GTCTGTCATACTCTAGCAGCACGTAAATATTGGTGTTAAAACTGTAAATATC TCCAGTATTA[A/-]CTGTGCTGCTGAAGTAAGGCT | 0.6 |
| MIR* | gga-mir-1648 | rs14281066 | G/A | TGGCCGGCGGCTCGGCTCGGCTCCGCTCACACCAGGAGCAGCGGCTCGGCGTGGTGGGAGTGGA[G/A]CGGAGCTGTCCGCCGTAGCCG | 0.6 |
| MIR* | gga-mir-1719 | rs14969646 | C/G | TTGAGAAGTACTTGTTTGTACCAAAAGATGATGAGTTGCTGATGTGCTTTGTCGAGGTAACGCT[G/C]TTTCCTGGGTGCAGTATCGCTTAA | 0.6 |
| MIR* | gga-mir-1596 | rs15190357 | C/T | TGGACTTTGTTTCATTCTCCTTCAATCCATCTTTGTCATGATTTACAAAAAG AACAAAGGTGGCC[G/A]TGGGTGAATGAAGCCAGGCATCCA | 0.5 |
| MIR* | gga-mir-1558 | rs14268122 | A/G | CGCTGCTCCGAGCCCGTTATA[T/C]TCAGCAGGACAAGAGGGGACAGGCAGGGCTGCTCACTGCTGTGATGGGAGCTCTGGGATGCG | 0.2 |
| MIR* | gga-mir-1605 | rs15600727 | T/C | AAGAGAAGTTCACAGAGGGTTTCATGTGTATCTCTTGAAGATGCTGATATG AGAGA[A/G]GCATATGAAAGCGCTTCTGTGATTTTTCTT | 0 |
| MIR* | gga-mir-1606 | rs14431112 | C/G | TGTGAGGCTGCTCTT[G/C]GGTTTCCCTCTGAAATTCACTGAAAGAGGGGGCAACAGAACAGTCCTGAACA | 0 |
| MIR* | gga-mir-1618 | rs14639002 | C/G | CTGCGTGCTGCATCCTGGCTCCTGG[C/G]GTCCTGCACGTGCTATTGGAGAGCCCGGAGCCAGGCTGCTGTTCTCCAGGCAG | 0 |
| MIR* | gga-mir-1630 | rs16651110 | T/C | TTTGGGCTTT[C/T]GTTTCTTCTTTCTAAAGGGGGAGGAAAGAGGAAACTGAAGCTGAAA | 0 |
| MIR* | gga-mir-1658 | rs16681032 | C/T | CCCCCTTCCTGAATATACCACCCCCAGGAGTTCTGCCATACCAGTGTGTGTTTCTCAGA[G/A]CTGTGGGTTGGTGTTGATGGGGAGCTGGGG | 0 |
| Other | gga-mir-1600 | rs14726712 | T/A | GCCCTTCTGGCTCCAGTCTTAACT[A/G]CAGGCAATGCTGACATTCTCCTTCAGTTATCAGTGGAGTTTGGAGTGC | 4.6 |
| Other | gga-mir-1768 | rs13538425 | T/G | TATCAGCTCATCAGCATGACAATCCTCTGCTGCCTTGGGAGTTGTGAGCAGAGGATTGTCGTGCTGA[T/G]GAGTTGATA | 4.3 |
| Other | gga-mir-1562 | rs14653234 | A/T | ATCAACAAGCGGGGTGCAGCTGCACTGTAGATGGAGGGGCTGCATGAAGGTGGATAGTTTCTGTGCCCCGTGTATGCAGTGCAGGTGCTC[T/T]CTGCACTT GGT | 4.1 |
| Other | gga-mir-1453 | rs16159272 | G/A | GTGCAGGCTCTCTGTGGCTCAGCCCAGCTG[T/C]GGTACTGCATGGTGCTGGGCAGCCTCTCAGTGAGCTCTGTGCT | 3.8 |
| 定位 Location | 基因 microRNA | SNP 登录号 dbSNP | 变异 Variation | Pre-microRNA序列变异 Variation of the pre-microRNA sequence | 自由能改变值 |
| ΔΔG (kcal/mol) | |||||
| Other | gga-mir-1666 | rs14120863 | C/G | GCCAGGAGCAAACACGA[G/C]TTAACGCCACGGGGCTGAGGCTGAGCTGCA AACTTTCACAGCTTCAGAGCGTGGCTGCAGCGCGTGCTGGC | 3.5 |
| Other | gga-mir-1704 | rs14668705 | C/G | GACCGA[C/G]AATTCACAGCAGATTGATGGGGCACACCAAGATCTGTGCCT GTTCCTGCTGGGATGCTCCCGTC | 3.4 |
| Other | gga-mir-1792 | rs14317161 | A/T | TCCAAAGCCACCTCTATGCTGCGATAGCTCAGGAACACTGCAGTCAAGAACACATACTGGAGTGTTCCTGGGGTATCACAGCGTAGGGGTGGT[T/A]TTGGA | 3.2 |
| Other | gga-mir-1812 | rs16571431 | A/G | CTTTATTGGGAACCTAGATATATACTGTATACTGTGTCAAGGTTTAGATGACATACTATACAGTATATATCTAGGTT[T/C]CTAATACAAG | 2.9 |
| Other | gga-mir-1608 | rs16662849 | A/G | TCTCATCTGGTGCCCCCGGGATGGGACAGTGGCTGCGCCCTCTGTGAAGGGAA[C/T]GGAGGGCTCAGCAGCTGTGCCCGTGCCTGGCAGAGTGGA | 2.5 |
| Other | gga-mir-1705 | rs15027520 | A/G | GCTGAGAGCTTTCCTCAACTGTGTGCTGATTTCATTAGACTTTTTCCTGACAAGAAATCTGGAAGTCAGCACATGCTGAGAAGA[G/A]TCTGC | 2.1 |
| Other | gga-mir-1757 | rs15505339 | G/C | GCTGTTTTGCCGTGGATGAACAAAACTCTTCTGGTCCGCTGTTTAGTTCAATTAGCGCTTCAGAGGCGGAACAGTAGGTTTCTGTTCTT[C/G]CAGCCAAGATTCTGT | 1.8 |
| Other | gga-mir-1749 | rs15860000 | C/T | GAGTGCAGCTGGAGGGCTACAGGGAGATAAGGAACTGAACTGGCTGCATCTCTGCTGCTTGGCTCTGTTCCCTATTTCC[C/T]GTCTCCTTAGCAAATCTC | 1.5 |
| Other | gga-mir-1657 | rs14934923 | A/G | GAGAAGTAT[A/G]TGTGTATACATCAACACATACTTATATTTGTATATGGAT TTACATATATGTGTATTGTTGGTGTGTGATACGATATTCCATATC | 1.2 |
| Other | gga-mir-1562 | rs14653232 | A/G | ACAACA[A/G]GCGGGGTGCAGCTGCACTGTAGATGGAGGGGCTGCATGAAGGTGGATAGTTTCTGTGCCCCGTGTATGCAGTGCAGGTGCTCACTGCACTGGT | 1.1 |
| Other | gga-mir-1704 | rs14668704 | C/T | GACCGAGAATTCACAGCAGATTGATGGGGCACACCAAGATCTGTGCCTGTTCCTGCTGGGATGCTCCC[G/A]TC | 1.0 |
| Other | gga-mir-1558 | rs15169308 | G/A | CGCTGCTCCAGAGCCCGTTATACTCAGCAGGACAAGAGGGGACAGGCAGGGCTGCTCACTGCTGTGATGGGAGCTCTGGGATG[C/T]G | 0.9 |
| Other | gga-mir-1790 | rs16361960 | G/A | GGCGGGGCGGGATGCCGCTGGGGC[T/C]GGGCCTCGTGCCGTGCTCTGCTGGGCCGGAGAAGGCGGGTGAGGGCGGTGGGGGAAGGGAAGCGGAGCTGTTTCTTCGCT | 0.8 |
| Other | gga-mir-1675 | rs16643638 | C/T | GCCTTCCC[G/A]CTTGGCCCCGCTCCCGCTCAGGGCGGTGTCAGGCGGCGCCGCCGGACGCCATCTTGACTGTGGGCAGCTCGGCTTCAGGTCGGCAGAGAGC | 0.7 |
| Other | gga-mir-551 | rs16677358 | A/G | CCACCAGAGC[T/G]GCCCCATGGCTCCAGAAATCAAGGGTGGGTAAGACCT TGTAGGATAACTGGCAGGCGACCCATACTTGGTTTCAGGGGCTGTGTGGGTATCG- TCAGTGGTGG | 0.6 |
| Other | gga-mir-1762 | rs13608151 | T/C | GGTGCTCCTCCAGCTGCAGGGCAGGAGGGAAGCAGAGCTTAGAGCTGTTTTCTACGCTCGAGCTGGCTTTGCTCAAGCCCCGTGTGCGG[A/G]GGGTTTCC | 0.5 |
| Other | gga-mir-1608 | rs15961488 | A/C | TCTCATCTGGTGCCCCCGGGATGGGACAGTGGCTGCGCCCTCTGTGAAGGGAATGGAGGGCTCAGCAGCTGTGCCCGTGCCTGGCAGAG[T/G]GGA | 0.5 |
| Other | gga-mir-1719 | rs14969645 | A/G | TTGAGAAGTACTTGTTTGTACCAAAAGATGATGAGTTGCTGATGTGCTTTGTCGAGGTAACGCTGTTTCCTGGG[T/C]GCAGTATCGCTTA | 0.4 |
| Other | gga-mir-1458 | rs16654946 | A/- | TTAGTTCAGCCTCA[T/-]TTTTTTTTAGAGTTCTGGAGTATCTTATTGCACAAAATATGCTGAGGAGAAATTCCTGTGATGCTCATGAGAAAAATTGTAGAGCGCTTTTACACT | 0.4 |
| Other | gga-mir-1562 | rs14653233 | G/A | ACAACAAGCGGG[G/A]TGCAGCTGCACTGTAGATGGAGGGGCTGCATGAAGGTGGATAGTTTCTGTGCCCCGTGTATGCAGTGCAGGTGCTCACTGCACTTGG GT | 0.4 |
| Other | gga-mir-1619 | rs14280616 | G/T | TTGTAAGTCCAGCAGTCTGTATTGCTACAAATGCTCTTCTCAGCGCAA[C/A] CTCAGAAGAATTCATAGCAATACGGACTGCTGGACTTCTAG | 0.3 |
| Other | gga-mir-1675 | rs16643636 | A/T | GCCTTCCCGCTTGGCCCCGCTCCCGCTCAGGGCGGTGTCAGGCGGCGCCG- CCGGACGCCATCTTGACTGTGGGCAGC[T/A]CGGCTTCAGGTCGGCAGAG- AGC | 0.3 |
| Other | gga-mir-1687 | rs15179830 | T/G | GATTTCTTTGTGTCTGTGGTCTGAGGAAATGAGCCAGCTGAGTTAGCAACTTCTTTGCTGGCTGTTTCTCAGTGACTGCAGCATAAAAA[T/G]C | 0.2 |
| Other | gga-mir-22 | rs14120771 | C/T | GCTCCTGGCTGCCC[G/A]GCAGCAGTTCTTCAGTGGCAAGCTTTATGTCCTTCTCTAGTAGCTAAAGCTGCCAGTTGAAGAACTGTTGAATGTAGCCACGTTC | 0 |
| Other | gga-mir-1659 | rs15847007 | G/A | CATAGTTTCATAT[C/T]CAGAGGAAGACTTCATTTTATTTAATGTTCTCATTTCACTGAATAGGGCAGAATGAAGGAG TCC CTGCCTGTGGTGAGTGACAGTG | 0 |
| Other | gga-mir-1590 | rs15781755 | A/G | TGGA[T/C]GGAAATAGGTATCTCCAGAGAACAGTTTGTCCCACTTGTTTTTGACGTCTGTCGTAGCAAGGGACAAATTGCTCTCTGGAAGTACATTTCTCCA | 0 |
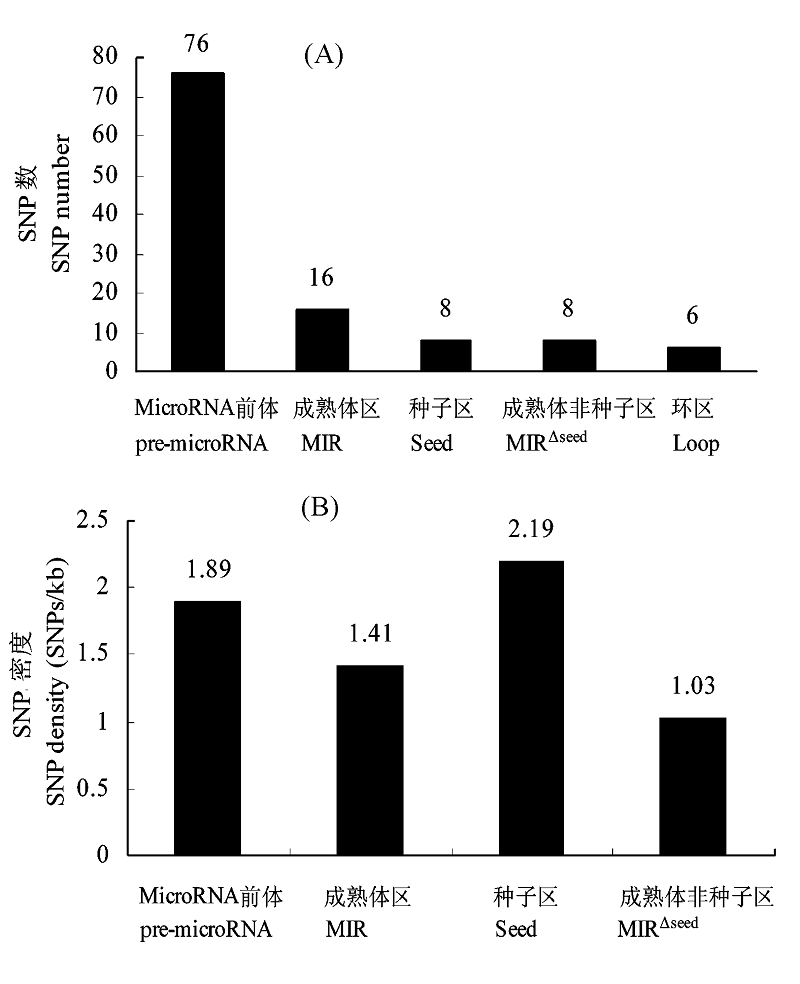
图3 SNP在pre-microRNAs二级结构的定位。(A) SNP在pre-microRNAs二级结构的分布; (B) pre-microRNAs二级结构不同区的SNP密度。
Fig. 3 Localization of SNP in pre-microRNA secondary structures. (A) Distribution of chicken SNP across the secondary structure of pre-microRNAs; (B) SNP density in different domains of the secondary structure of pre-microRNAs.
| 定位 Location | 个数 Number | 均值 ± 标准误 Mean ± SE | 最小值 Minimum | 最大值 Maximum |
|---|---|---|---|---|
| Loop | 6 | 0.5667 ± 0.34897 a | 0.00 | 1.90 |
| MIRΔseed | 8 | 1.6750 ± 0.57001 ab | 0.30 | 4.00 |
| Seed | 8 | 3.4500 ± 0.92157 b | 0.00 | 7.00 |
| MIR* | 24 | 1.7208 ± 0.37003 a | 0.00 | 6.20 |
| Other | 30 | 1.5667 ± 0.26916 a | 0.00 | 4.60 |
表2 Pre-microRNA二级结构不同区域SNP的ΔΔG均值的多重比较(LSD)
Table 2 Multiple range comparison between the mean value of ΔΔG of the SNP located in different domains of pre-microRNAs,secondary structures
| 定位 Location | 个数 Number | 均值 ± 标准误 Mean ± SE | 最小值 Minimum | 最大值 Maximum |
|---|---|---|---|---|
| Loop | 6 | 0.5667 ± 0.34897 a | 0.00 | 1.90 |
| MIRΔseed | 8 | 1.6750 ± 0.57001 ab | 0.30 | 4.00 |
| Seed | 8 | 3.4500 ± 0.92157 b | 0.00 | 7.00 |
| MIR* | 24 | 1.7208 ± 0.37003 a | 0.00 | 6.20 |
| Other | 30 | 1.5667 ± 0.26916 a | 0.00 | 4.60 |
| 靶基因 Target gene | 突变体 Mutation type | 野生型 Wild type | 吉布斯自由能改变量 ΔΔG (kcal/mol) |
|---|---|---|---|
| DOCK9 | 3' GCT-AC-A-T-C-A-G-GT-TCCCGTGTAT 5'(-14.60) | | || | | : :| | |||||||| 5' C-AGTGTTCTTTTTTTGTGTATGGCACATA 3' | 14.60 | |
| TNRC6B | 3' GCT-ACATCAGGTTCCCGTGTAT 5'(-19.36) | | ||| ||:| |||||||| 5' C-AGTGT-GTTC-T-GGCACATG 3' | 19.36 | |
| CPEB1 | 3' GCTACATCAGGTTCCCCGTGTAT 5' (-26.26) :|:||||||:: |||||||| 5' TGGTGTAGTTTT-T-GGCACATA 3' | 3' GCTACATCAGGTTCCCGTGTAT 5'(-26.94) :|:||||||:: |||||||| 5' TGGTGTAGTTT-TTGGCACATA 3' | 0.68 |
| KCNJ2 | 3' GCTAC - A - TCA - G - GT -TC - CCCGTGTAT 5'(-18.7) | ||| | ||| : | || |||||||| 5' C-ATGTCTAAGTATGAACAGAAGGCACATA 3' | 3' GCTACAT-C-AG-GTTCCCGTGTAT 5'(-16.67) | | ||| | | || |||||||| 5' CTAAGTATGAACAGAA-GGCACATA 3' | 2.03 |
| WDR22 | 3' GC-TACATCA-GGTTCCCCGTGTAT 5'(-17.3) || |||| || ::|| |||||||| 5' CGTAGTGTGTATTAA-AGGCACATA 3' | 3' GC-T-ACATCA-GGTTCCCGTGTAT 5'(-18.37) || | ||||| ::|| |||||||| 5' CGTAGTGT-GTATTAAAGGCACATA 3' | 1.07 |
| ADD3 | 3' GCTACATCAGGT - T-CCCGTGTAT 5'(-14.65) || || |::| | |||||||| 5' AGAAATATTATACACTGGCACATA 3' | 14.65 | |
| PAPOLG | 3' GCTACAT-CAGGTTC-C-CCGTGTAT 5'(-19.88) | ||| ||| || |||||||| 5' AAA-GTATTTCC-AGTTTGGCACATA 3' | 3' GCTACAT-CAGGTT - C-CCGTGTAT 5'(-19.49) | ||| ||||| |||||||| 5' AAA-GTATTTCCAGTTTGGCACATA 3' | 0.39 |
| PPP1R12A | 3' GCTACATCAGGTTCCCCGTGTAT 5'(-16.71) ||| | ||| | |||||||| 5' CGA-C-A-TCCTATCGGCACATA 3' | 3' GCTACATCAGGTT-CCCGTGTAT 5'(-16.32) ||| | ||| | ||||||| 5' CGA-C-A-TCCTATCGGCACATA 3' | 0.39 |
表3 SNP多态对gga-mir-455-5p靶基因选择的影响。括号内显示的为吉布斯自由能值。
Table 3 Effect of the SNP polymorphism on the target gene of gga-mir-455-5p. Gibbs free-energy are shown in brackets.
| 靶基因 Target gene | 突变体 Mutation type | 野生型 Wild type | 吉布斯自由能改变量 ΔΔG (kcal/mol) |
|---|---|---|---|
| DOCK9 | 3' GCT-AC-A-T-C-A-G-GT-TCCCGTGTAT 5'(-14.60) | | || | | : :| | |||||||| 5' C-AGTGTTCTTTTTTTGTGTATGGCACATA 3' | 14.60 | |
| TNRC6B | 3' GCT-ACATCAGGTTCCCGTGTAT 5'(-19.36) | | ||| ||:| |||||||| 5' C-AGTGT-GTTC-T-GGCACATG 3' | 19.36 | |
| CPEB1 | 3' GCTACATCAGGTTCCCCGTGTAT 5' (-26.26) :|:||||||:: |||||||| 5' TGGTGTAGTTTT-T-GGCACATA 3' | 3' GCTACATCAGGTTCCCGTGTAT 5'(-26.94) :|:||||||:: |||||||| 5' TGGTGTAGTTT-TTGGCACATA 3' | 0.68 |
| KCNJ2 | 3' GCTAC - A - TCA - G - GT -TC - CCCGTGTAT 5'(-18.7) | ||| | ||| : | || |||||||| 5' C-ATGTCTAAGTATGAACAGAAGGCACATA 3' | 3' GCTACAT-C-AG-GTTCCCGTGTAT 5'(-16.67) | | ||| | | || |||||||| 5' CTAAGTATGAACAGAA-GGCACATA 3' | 2.03 |
| WDR22 | 3' GC-TACATCA-GGTTCCCCGTGTAT 5'(-17.3) || |||| || ::|| |||||||| 5' CGTAGTGTGTATTAA-AGGCACATA 3' | 3' GC-T-ACATCA-GGTTCCCGTGTAT 5'(-18.37) || | ||||| ::|| |||||||| 5' CGTAGTGT-GTATTAAAGGCACATA 3' | 1.07 |
| ADD3 | 3' GCTACATCAGGT - T-CCCGTGTAT 5'(-14.65) || || |::| | |||||||| 5' AGAAATATTATACACTGGCACATA 3' | 14.65 | |
| PAPOLG | 3' GCTACAT-CAGGTTC-C-CCGTGTAT 5'(-19.88) | ||| ||| || |||||||| 5' AAA-GTATTTCC-AGTTTGGCACATA 3' | 3' GCTACAT-CAGGTT - C-CCGTGTAT 5'(-19.49) | ||| ||||| |||||||| 5' AAA-GTATTTCCAGTTTGGCACATA 3' | 0.39 |
| PPP1R12A | 3' GCTACATCAGGTTCCCCGTGTAT 5'(-16.71) ||| | ||| | |||||||| 5' CGA-C-A-TCCTATCGGCACATA 3' | 3' GCTACATCAGGTT-CCCGTGTAT 5'(-16.32) ||| | ||| | ||||||| 5' CGA-C-A-TCCTATCGGCACATA 3' | 0.39 |
| [1] |
Bennett AK, Hester PY, Spurlock DE (2007) Polymorphisms in vitamin D receptor, osteopontin, insulin-like growth factor 1 and insulin, and their associations with bone, egg and growth traits in a layer-broiler cross in chickens. Animal Genetics, 37,283-286.
DOI URL PMID |
| [2] | Borel C, Antonarakis SE (2008) Functional genetic variation of human miRNAs and phenotypic consequences. Mammalian Genome, 19(7-8),503-509. |
| [3] |
Duan R, Pak C, Jin P (2007) Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Human Molecular Genetics, 16,1124-1131.
URL PMID |
| [4] |
Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS (2003) MicroRNA targets in Drosophila. Genome Biology, 5,R1.
URL PMID |
| [5] | Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML (2008) A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Research, 18,957-964. |
| [6] | Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Research, 36,D154-D158. |
| [7] |
Hicks JA, Tembhurne P, Liu HC (2008) MicroRNA expression in chicken embryos. Poultry Science, 87,2335-2343.
DOI URL PMID |
| [8] | Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle (2008) A common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proceedings of the National Academy of SciencesUSA, 105,7269-7274. |
| [9] | Landi D, Gemignani F, Barale R, Landi S (2008) A catalog of polymorphisms falling in microrna-binding regions of cancer genes. DNA and Cell Biology, 27,35-43. |
| [10] |
Lei M, Luo C, Peng X, Fang M, Nie QH, Zhang D, Yang GF, Zhang XQ (2007) Polymorphism of growth-correlated genes associated with fatness and muscle fiber traits in chickens. Poultry Science, 86,835-842.
DOI URL PMID |
| [11] | Min LJ (闵令江), Pan QJ (潘庆杰), Chen H (陈宏), Lei CC (雷初朝), Li MY (李美玉), Sun GQ (孙国强) (2005) Studies of insulin-like growth factor-I ( IGF-I) gene polymorphism analysis and its associations with body weight and carcass traits in Shouguang chicken. Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 36,645-648. (in Chinese with English abstract) |
| [12] |
Mishra PJ, Mishra PJ, Banerjee D, Bertino JR (2008) MiRSNP or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle, 7,853-858.
URL PMID |
| [13] | Rao YS (饶友生), Zhang XQ (张细权) (2007) Single nucleotide polymorphisms and fine mapping QTL in chickens. Hereditas (Beijing)(遗传), 29,393-398. (in Chinese with English abstract) |
| [14] | Saunders MA, Liang H, Li WH (2007) Human polymorphism at microRNAs and microRNA target sites. Proceedings of the National Academy of Sciences, USA, 104,3300-3305. |
| [15] | Wang J, He XM, Ruan J, Dai MT, Chen J, Zhang Y, Hu YF, Chen Y, Li ST, Cong LJ, Fang L, Liu B, Li SG, Wang J, Burt DW, Wong GK, Yu J, Yang HM, Wang J (2005) ChickVD: a sequence variation database for the chicken genome. Nucleic Acids Research, 33,D438-441. |
| [16] | Wu X (吴旭), Wang JY (王金玉), Yan MJ (严美姣), Li HF (李慧芳), Chen KW (陈宽维), Tang QP (汤青萍), Zhu WQ (朱文奇), Yu YB (俞亚波) (2007) Genetic effects of GNRHR and IGF-I genes on the reproductive traits in Wenchang chicken. Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 38,31-35. (in Chinese with English abstract) |
| [17] |
Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, Su H, Zhuang SM (2008) A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis, 29,2126-2131.
URL PMID |
| [18] |
Zeng Y, Cullen BR (2003) Sequence requirements for microRNA processing and function in human cells. RNA, 9,112-123.
DOI URL PMID |
| [19] | Zhang QP, Lu M, Cui QG (2008) SNP analysis reveals an evolutionary acceleration of the human-specific microRNAs. Nature Precedings. http://precedings.nature.com/documents/2127/version/1/html. |
| [20] | Zhu Z (朱智), Xu NY (徐宁迎), Wu DJ (吴登俊), Huang LQ (黄利权), Zhao XF (赵晓枫), Zhang XY (张翔宇) (2007) SNP of IGF-I gene and its genetic effects on carcass traits in chicken. Acta Veterinaria et Zootechnica Sinica (畜牧兽医学报), 38,1021-1026. (in Chinese with English abstract) |
| [1] | 徐聪, 张飞宇, 俞道远, 孙新, 张峰. 土壤动物的分子分类预测策略评估[J]. 生物多样性, 2022, 30(12): 22252-. |
| [2] | 莫日根高娃, 商辉, 刘保东, 康明, 严岳鸿. 一个种还是多个种? 简化基因组及其形态学证据揭示中国白桫椤植物的物种多样性分化[J]. 生物多样性, 2019, 27(11): 1196-1204. |
| [3] | 徐晓婷, 王志恒, DimitarDimitrov. 批量下载GenBank基因序列数据的新工具——NCBIminer[J]. 生物多样性, 2015, 23(4): 550-555. |
| [4] | 孙欣, 高莹, 杨云锋. 环境微生物的宏基因组学研究新进展[J]. 生物多样性, 2013, 21(4): 393-400. |
| [5] | 邹喻苹, 葛颂. 新一代分子标记——SNPs及其应用[J]. 生物多样性, 2003, 11(5): 370-382. |
| [6] | 钟扬, 张亮, 任文伟, 陈家宽. 生物多样性信息学:一个正在兴起的新方向及其关键技术[J]. 生物多样性, 2000, 08(4): 397-404. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
备案号:京ICP备16067583号-7
Copyright © 2022 版权所有 《生物多样性》编辑部
地址: 北京香山南辛村20号, 邮编:100093
电话: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn