



Biodiv Sci ›› 2010, Vol. 18 ›› Issue (3): 227-232. DOI: 10.3724/SP.J.1003.2010.227 cstr: 32101.14.SP.J.1003.2010.227
• Special Issue • Previous Articles Next Articles
Received:2009-10-19
Accepted:2009-12-16
Online:2010-05-20
Published:2012-02-08
Contact:
Daming Zhang
Yingying Cui, Daming Zhang. Surveying DNA methylation diversity in the wild rice, Oryza nivaraand O. rufipogon[J]. Biodiv Sci, 2010, 18(3): 227-232.
| 序号 Index | IRGC编号 IRGC No. | 来源地 Geographic origin | ||||||
|---|---|---|---|---|---|---|---|---|
| O. nivara | ||||||||
| N1 | 103407 | 斯里兰卡 Sri LanKa | ||||||
| N2 | 105431 | 斯里兰卡 Sri LanKa | ||||||
| N3 | 105391 | 泰国 Thailand | ||||||
| N4 | 106345 | 缅甸 Myanmar | ||||||
| N5 | 106185 | 印度 India | ||||||
| O. rufipogon | ||||||||
| R1 | 106515 | 越南 Vietnam | ||||||
| R2 | 82000 | 巴布亚新几内亚 Papua New Guinea | ||||||
| R3 | 103423 | 斯里兰卡 Sri LanKa | ||||||
| R4 | 105958 | 印度尼西亚 Indonesia | ||||||
| R5 | 106036 | 马来西亚 Malaysia | ||||||
Table 1 Oryza nivaraand O. rufipogon accessions studied
| 序号 Index | IRGC编号 IRGC No. | 来源地 Geographic origin | ||||||
|---|---|---|---|---|---|---|---|---|
| O. nivara | ||||||||
| N1 | 103407 | 斯里兰卡 Sri LanKa | ||||||
| N2 | 105431 | 斯里兰卡 Sri LanKa | ||||||
| N3 | 105391 | 泰国 Thailand | ||||||
| N4 | 106345 | 缅甸 Myanmar | ||||||
| N5 | 106185 | 印度 India | ||||||
| O. rufipogon | ||||||||
| R1 | 106515 | 越南 Vietnam | ||||||
| R2 | 82000 | 巴布亚新几内亚 Papua New Guinea | ||||||
| R3 | 103423 | 斯里兰卡 Sri LanKa | ||||||
| R4 | 105958 | 印度尼西亚 Indonesia | ||||||
| R5 | 106036 | 马来西亚 Malaysia | ||||||
| 引物与接头 Primer and adapter | 序列(5′-3′) Sequence(5′-3′) |
|---|---|
| 接头 Adapter | |
| EcoRI-adapter I | CTGGTAGACTGCGTACC |
| EcoRI-adapter II | AATTGGTACGCAGTC |
| 预扩增引物 Preselective primer | |
| EcoRI + A | GACTGCGTACCAATTCA |
| EcoRI + C | GACTGCGTACCAATTCC |
| 选择性扩增引物 Selective primer | |
| EcoRI + AA | GACTGCGTACCAATTCAA |
| EcoRI + AT | GACTGCGTACCAATTCAT |
| EcoRI + AC | GACTGCGTACCAATTCAC |
| EcoRI + AG | GACTGCGTACCAATTCAG |
| EcoRI + CT | GACTGCGTACCAATTCCT |
| EcoRI + CG | GACTGCGTACCAATTCCG |
| EcoRI + CA | GACTGCGTACCAATTCCA |
Table 2 Sequences of adapter and primer used for MSAP analysis
| 引物与接头 Primer and adapter | 序列(5′-3′) Sequence(5′-3′) |
|---|---|
| 接头 Adapter | |
| EcoRI-adapter I | CTGGTAGACTGCGTACC |
| EcoRI-adapter II | AATTGGTACGCAGTC |
| 预扩增引物 Preselective primer | |
| EcoRI + A | GACTGCGTACCAATTCA |
| EcoRI + C | GACTGCGTACCAATTCC |
| 选择性扩增引物 Selective primer | |
| EcoRI + AA | GACTGCGTACCAATTCAA |
| EcoRI + AT | GACTGCGTACCAATTCAT |
| EcoRI + AC | GACTGCGTACCAATTCAC |
| EcoRI + AG | GACTGCGTACCAATTCAG |
| EcoRI + CT | GACTGCGTACCAATTCCT |
| EcoRI + CG | GACTGCGTACCAATTCCG |
| EcoRI + CA | GACTGCGTACCAATTCCA |
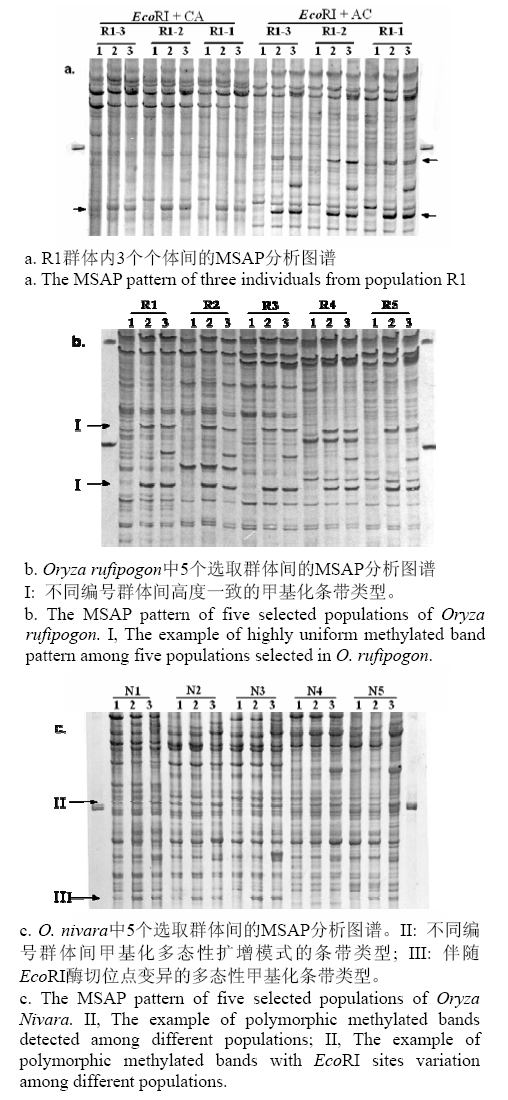
Fig. 1 Part of electrophoretic profiles obtained using MSAP in the wild rice, Oryza nivara and O. Rufipogon.Lanes 1-3 show three digestion and amplification production (1, EcoRI / MspI; 2, EcoRI /HpaII and 3:EcoRI), respectively. The arrowheads indicate fragments containing the methylated CCGG sites.
| 种 Species | 大小 Size (bp) | E值 E value | 基因库 GenBank | 染色体 Chromosome | 检索结果 Result of BLAST |
|---|---|---|---|---|---|
| Class I | |||||
| O. nivara | 494 | 0 | AP008217 | 11,12 | Internal region of two genes; predicted as promoter region (0.9) |
| 644 | 0 | AP008217 | 11,12 | Internal region of two genes; predicted as promoter region (0.9) | |
| O. rufipogon | 615 | 0 | AP005531 | 8 | RNA-binding region RNP-1containing protein (BAD05783) |
| 640 | 0 | AP008218 | 11,12 | Internal region of two genes; predicted as promoter region (0.85) | |
| 548 | 0 | AP008212 | 6 | Putative mitochondrial phosphate transporter (BAD35704.1) | |
| 282 | 6.00E-13 | AC104322 | 3 | Putative polyprotein (AAR89842.1) | |
| Class II | |||||
| O. nivara | 742 | 0 | AP008216.1 | 2,10 | Internal region of two genes; predicted as promoter region (0.97/ 0.89) |
| 630 | 0 | AP005563 | 9 | TPR-like domain containing protein (BAD19882.1) | |
| 218 | 4.00E-61 | AL731583 | 4 | Ribonuclease HI (CAE 05186.2) | |
| O. rufipogon | 366 | 0 | AP008210 | 4 | Kinesin, motor region domain containing protein (CAE04590.2) |
| 563 | 7.00E-56 | AP008213 | 7 | Internal region of two genes; predicted as promoter region (0.99) | |
| Class III | |||||
| O. nivara | 569 | 0 | AC120529 | 3 | Near the centromere, the internal region of putative protein and retrotransposon |
| 305 | 2.00E-11 | AP008208.1 | 2 | Inetrnal region of a pseudo gene | |
| 681 | 0 | AP008209 | 3 | Snabo-1 miniature inverted repeat transposable element/ the intron of pseudo gene | |
| 615 | 0 | AC120529 | 3 | Near the centromere, the internal region of putative protein and retrotransposon | |
| O. rufipogon | 478 | 0 | AC120529 | 3 | Near the centromere, the internal region of putative protein and retrotransposon |
| 599 | 0 | AP004364.4 | 11,12 | Internal region of two hypotetical protein | |
| 514 | 0 | AP008209.1 | 3 | Near the centromere, the internal region of putative protein and retrotransposon |
Table 3 BLAST (short of The Basic Local Alignment Search Tool) search results for sequenced methylated fragments
| 种 Species | 大小 Size (bp) | E值 E value | 基因库 GenBank | 染色体 Chromosome | 检索结果 Result of BLAST |
|---|---|---|---|---|---|
| Class I | |||||
| O. nivara | 494 | 0 | AP008217 | 11,12 | Internal region of two genes; predicted as promoter region (0.9) |
| 644 | 0 | AP008217 | 11,12 | Internal region of two genes; predicted as promoter region (0.9) | |
| O. rufipogon | 615 | 0 | AP005531 | 8 | RNA-binding region RNP-1containing protein (BAD05783) |
| 640 | 0 | AP008218 | 11,12 | Internal region of two genes; predicted as promoter region (0.85) | |
| 548 | 0 | AP008212 | 6 | Putative mitochondrial phosphate transporter (BAD35704.1) | |
| 282 | 6.00E-13 | AC104322 | 3 | Putative polyprotein (AAR89842.1) | |
| Class II | |||||
| O. nivara | 742 | 0 | AP008216.1 | 2,10 | Internal region of two genes; predicted as promoter region (0.97/ 0.89) |
| 630 | 0 | AP005563 | 9 | TPR-like domain containing protein (BAD19882.1) | |
| 218 | 4.00E-61 | AL731583 | 4 | Ribonuclease HI (CAE 05186.2) | |
| O. rufipogon | 366 | 0 | AP008210 | 4 | Kinesin, motor region domain containing protein (CAE04590.2) |
| 563 | 7.00E-56 | AP008213 | 7 | Internal region of two genes; predicted as promoter region (0.99) | |
| Class III | |||||
| O. nivara | 569 | 0 | AC120529 | 3 | Near the centromere, the internal region of putative protein and retrotransposon |
| 305 | 2.00E-11 | AP008208.1 | 2 | Inetrnal region of a pseudo gene | |
| 681 | 0 | AP008209 | 3 | Snabo-1 miniature inverted repeat transposable element/ the intron of pseudo gene | |
| 615 | 0 | AC120529 | 3 | Near the centromere, the internal region of putative protein and retrotransposon | |
| O. rufipogon | 478 | 0 | AC120529 | 3 | Near the centromere, the internal region of putative protein and retrotransposon |
| 599 | 0 | AP004364.4 | 11,12 | Internal region of two hypotetical protein | |
| 514 | 0 | AP008209.1 | 3 | Near the centromere, the internal region of putative protein and retrotransposon |
| 类型 Class | 序列号 Accession no. | 注释 Note | 编码区长度 Coding region (bp) | 同源序列比对 Sequence contrast | 非同义突变位点 Non synonymy mutation sites | 同义突变位点 Synonymy mutation sites |
|---|---|---|---|---|---|---|
| Class I | Os08g0190200 | RNA recognition motif | 391 | rufipogon-japonica rufipogon-indica | 1 0 | 0 2 |
| Os06g0210500 | Putative mitochondrial phosphate transporter | 150 | rufipogon-japonica rufipogon-indica | 0 0 | 0 0 | |
| OSJNBa0024F18.68 | Putative polyprotein | 281 | rufipogon-japonica rufipogon-indica | 0 0 | 4 5 | |
| Class II | Os09g0124100 | TPR-like domain containing protein | 128 | nivara-japonica nivara-indica | 1 1 | 0 0 |
| OSJNBb0069N01.4 | Ribonuclease HI | 207 | nivara-japonica nivara-indica | 11 11 | 6 7 | |
| Os04g0375900 | Kinesin, motor region domain containing protein | 241 | rufipogon-japonica rufipogon-indica | 0 4 | 0 1 |
Table 4 Comparisons of partial exon sequences of six genes with the corresponding sequences of japonica and indica
| 类型 Class | 序列号 Accession no. | 注释 Note | 编码区长度 Coding region (bp) | 同源序列比对 Sequence contrast | 非同义突变位点 Non synonymy mutation sites | 同义突变位点 Synonymy mutation sites |
|---|---|---|---|---|---|---|
| Class I | Os08g0190200 | RNA recognition motif | 391 | rufipogon-japonica rufipogon-indica | 1 0 | 0 2 |
| Os06g0210500 | Putative mitochondrial phosphate transporter | 150 | rufipogon-japonica rufipogon-indica | 0 0 | 0 0 | |
| OSJNBa0024F18.68 | Putative polyprotein | 281 | rufipogon-japonica rufipogon-indica | 0 0 | 4 5 | |
| Class II | Os09g0124100 | TPR-like domain containing protein | 128 | nivara-japonica nivara-indica | 1 1 | 0 0 |
| OSJNBb0069N01.4 | Ribonuclease HI | 207 | nivara-japonica nivara-indica | 11 11 | 6 7 | |
| Os04g0375900 | Kinesin, motor region domain containing protein | 241 | rufipogon-japonica rufipogon-indica | 0 4 | 0 1 |
| [1] |
Alexandre CM, Hennig L (2008) FLC or not FLC: the other side of vernalization. Journal of Experimental Botany, 59, 1127-1135.
DOI URL PMID |
| [2] |
Ashikawa I (2001) Surveying CpG methylation at 5'-CCGG in the genomes of rice cultivars. Plant Molecular Biology, 45, 31-39.
DOI URL PMID |
| [3] |
Cervera MT, Ruiz-García L, Martínez-Zapater JM (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Molecular Genetics and Genomics, 268, 543-552.
URL PMID |
| [4] |
Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nature Reviews Genetics, 6, 351-360.
DOI URL PMID |
| [5] |
Cubas P, Vincent C, Coen E (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature, 401, 157-161.
URL PMID |
| [6] |
Gruenbaum Y, Naveh-Many T, Ceder H, Razin A (1981) Sequence specificity of methylation in higher plant DNA. Nature, 292, 860-862.
DOI URL PMID |
| [7] |
Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Molecular Biology, 43, 179-188.
DOI URL PMID |
| [8] | Keyte AL, Percififld R, Liu B, Wendel JF (2006) Intraspecific DNA methylation polymorphism in cotton ( Gossypim hir- sutum L.). Journal of Heredity, 97, 444-450. |
| [9] |
Knox MR, Ellis THN (2001) Stability and inheritance of methylation states at PstI sites in Pisum. Molecular Genetics and Genomics, 265, 497-507.
DOI URL PMID |
| [10] |
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 8, 4321-4325.
DOI URL PMID |
| [11] | Nelson M, McClelland M (1991) Site-specific methylation: effect on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Research, 19S, 2045-2071. |
| [12] |
Susan K, Michael DP (2004) Epialleles via DNA methylation: consequences for plant evolution. Trends in Ecology and Evolution, 19, 309-314.
DOI URL PMID |
| [13] | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 24, 4876-4882. |
| [14] |
Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff S (2005) DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Current Biology, 15, 154-159.
DOI URL PMID |
| [15] |
Wang YM, Lin XY, Dong B, Wang YD, Liu B (2004) DNA methylation polymorphism in a set of elite rice cultivars and its possible contribution to inter-cultivar differential gene expression. Cellular and Molecular Biology Letters, 9, 543-556.
URL PMID |
| [16] |
Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Q (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Molecular and General Genetics, 261, 439-446.
DOI URL PMID |
| [17] |
Zilberman D (2008) The evolving functions of DNA methylation. Current Opinion in Plant Biology, 11, 554-559.
DOI URL PMID |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Copyright © 2022 Biodiversity Science
Editorial Office of Biodiversity Science, 20 Nanxincun, Xiangshan, Beijing 100093, China
Tel: 010-62836137, 62836665 E-mail: biodiversity@ibcas.ac.cn ![]()
