Gene ontology: Tool for the unification of biology
1
2000
... 基于Ks分布结果, 调取峰值范围95%的基因, 对其进行功能注释(Ashburner et al, 2000).采用agriGO v2.0中的Fisher检验对保留下来的复制基因进行GO功能富集(中国农业大学, 北京) (P < 0.05) (Tian et al, 2017). ...
Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana
1
2000
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
The sunflower genome provides insights into oil metabolism, flowering and asterid evolution
1
2017
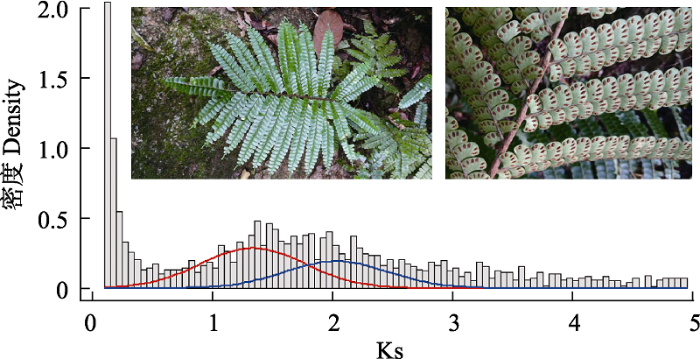
... 目前全基因组复制事件检测常用的方法是同义替换率(Ks)法.它不影响氨基酸的正常编码, 为中性突变(Kimura, 1977), 假定其以近似恒定的速率累积, Ks值在某种程度上可指示同源基因的发生时间.正常情况下, 物种的Ks分布图呈现L分布, 分布图前段的峰值反映的是近期的基因复制.多倍化发生后会产生大量同源基因, 映射到Ks分布图上便会有大量Ks值接近的同源基因对产生.如果在分布图中有明显其他峰值存在, 则表明在这个Ks值的时间段内有大量的同源基因产生, 即全基因组复制事件(Lynch & Conery, 2000; Vanneste et al, 2013).另外, 假定同义替换以恒定速率堆积, 可以根据峰值对应的Ks值估算全基因组复制事件发生的时间(Badouin et al, 2017).根据公式T = Ks/2r可以推断物种的分化时间, 本研究r选用水蕨(Ceratopteris thalictroides)的1.104 × 10-8计算复制发生时间(Zhang et al, 2019). ...
Evolutionary genomic analyses of ferns reveal that high chromosome numbers are a product of high retention and fewer rounds of polyploidy relative to angiosperms
1
2009
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
On the relative abundance of auto- and allopolyploids
1
2015
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes
1
2004
... 本研究将Ks值小于0.1以及大于5的结果去除, 以排除随机误差以及同义替换饱和效应的影响(Blanc & Wolfe, 2004; Schlueter et al, 2004; Cui et al, 2006).为了避免峰的假阳性, 运用mclust中高斯混合模型对数据进行正态分布拟合(Fraley & Raftery, 2003). ...
Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events
1
2003
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Origin of horsetails and the role of whole-genome duplication in plant macroevolution
1
2019
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
Widespread genome duplications throughout the history of flowering plants
2
2006
... 本研究将Ks值小于0.1以及大于5的结果去除, 以排除随机误差以及同义替换饱和效应的影响(Blanc & Wolfe, 2004; Schlueter et al, 2004; Cui et al, 2006).为了避免峰的假阳性, 运用mclust中高斯混合模型对数据进行正态分布拟合(Fraley & Raftery, 2003). ...
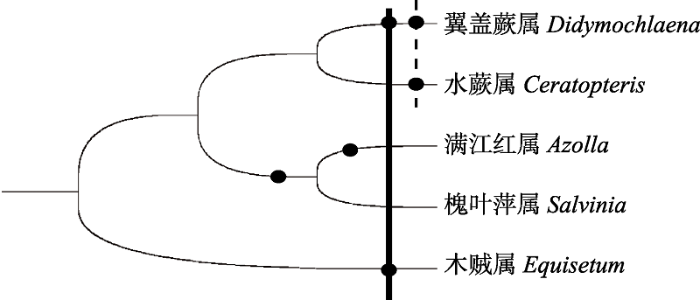
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...
Genome duplication and the origin of angiosperms
1
2005
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Genome size expansion and the relationship between nuclear DNA content and spore size in the Asplenium monanthes fern complex (Aspleniaceae)
1
2013
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
MUSCLE: Multiple sequence alignment with high accuracy and high throughput
1
2004
... 对物种内CDS序列进行all-against-all blast, 以e-5为阈值筛选基因家族中的旁系同源基因对, 使用mclblastline pipeline构建基因家族(Enright et al, 2002), 然后利用MUSCLE对每个基因家族进行比对(Edgar, 2004).运用PAML包中的CODEML软件计算得到Ks值(Goldman & Yang, 1994; Yang, 2007). ...
An efficient algorithm for large-scale detection of protein families
1
2002
... 对物种内CDS序列进行all-against-all blast, 以e-5为阈值筛选基因家族中的旁系同源基因对, 使用mclblastline pipeline构建基因家族(Enright et al, 2002), 然后利用MUSCLE对每个基因家族进行比对(Edgar, 2004).运用PAML包中的CODEML软件计算得到Ks值(Goldman & Yang, 1994; Yang, 2007). ...
Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proceedings of the National Academy of Sciences,
1
2009
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...
Enhanced model-based clustering, density estimation, and discriminant analysis software: MCLUST
1
2003
... 本研究将Ks值小于0.1以及大于5的结果去除, 以排除随机误差以及同义替换饱和效应的影响(Blanc & Wolfe, 2004; Schlueter et al, 2004; Cui et al, 2006).为了避免峰的假阳性, 运用mclust中高斯混合模型对数据进行正态分布拟合(Fraley & Raftery, 2003). ...
Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition
1
2009
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
A codon-based model of nucleotide substitution for protein-coding DNA sequences
1
1994
... 对物种内CDS序列进行all-against-all blast, 以e-5为阈值筛选基因家族中的旁系同源基因对, 使用mclblastline pipeline构建基因家族(Enright et al, 2002), 然后利用MUSCLE对每个基因家族进行比对(Edgar, 2004).运用PAML包中的CODEML软件计算得到Ks值(Goldman & Yang, 1994; Yang, 2007). ...
Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms
1
2015
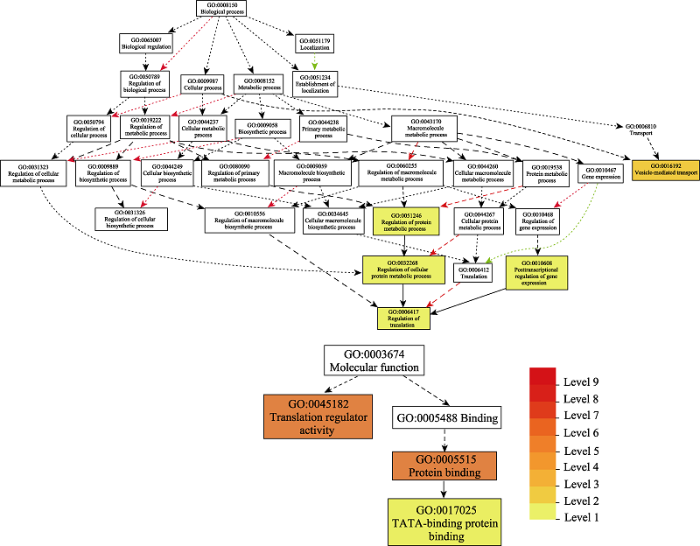
... 通过对保留下来的复制基因进行功能富集分析(图2), 发现调控蛋白代谢过程(GO:0051246)、调控胞内蛋白代谢过程(GO:0032268)、调控翻译(GO:0006417)、基因表达转录后调控(GO:0010608)及翻译调控活性(GO:0045182)等GO terms被优势保留, 翼盖蕨可能通过调控转录、转录后调控、影响翻译起始因子活性, 并通过调控蛋白代谢, 参与逆境胁迫调控网络, 从而增强翼盖蕨对极端环境的适应性(Muñoz & Castellano, 2012; Guerra et al, 2015); 此外, 还富集到TATA-binding protein binding (GO:0017025), 其主要功能是通过控制Transcription factor IIB-related protein, 影响植物的萌发率(Niu et al, 2013).推测这些复制基因的偏倚性保留对于翼盖蕨的环境适应性起重要作用. ...
Genomic clues to the evolutionary success of polyploid plants
1
2008
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Recurrent genome duplication events likely contributed to both the ancient and recent rise of ferns
1
2019
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics
1
2016
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Ancestral polyploidy in seed plants and angiosperms
2
2011
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
... ).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution
1
1977
... 目前全基因组复制事件检测常用的方法是同义替换率(Ks)法.它不影响氨基酸的正常编码, 为中性突变(Kimura, 1977), 假定其以近似恒定的速率累积, Ks值在某种程度上可指示同源基因的发生时间.正常情况下, 物种的Ks分布图呈现L分布, 分布图前段的峰值反映的是近期的基因复制.多倍化发生后会产生大量同源基因, 映射到Ks分布图上便会有大量Ks值接近的同源基因对产生.如果在分布图中有明显其他峰值存在, 则表明在这个Ks值的时间段内有大量的同源基因产生, 即全基因组复制事件(Lynch & Conery, 2000; Vanneste et al, 2013).另外, 假定同义替换以恒定速率堆积, 可以根据峰值对应的Ks值估算全基因组复制事件发生的时间(Badouin et al, 2017).根据公式T = Ks/2r可以推断物种的分化时间, 本研究r选用水蕨(Ceratopteris thalictroides)的1.104 × 10-8计算复制发生时间(Zhang et al, 2019). ...
Fern genomes elucidate land plant evolution and cyanobacterial symbioses
1
2018
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
The evolutionary fate and consequences of duplicate genes
2
2000
... 目前全基因组复制事件检测常用的方法是同义替换率(Ks)法.它不影响氨基酸的正常编码, 为中性突变(Kimura, 1977), 假定其以近似恒定的速率累积, Ks值在某种程度上可指示同源基因的发生时间.正常情况下, 物种的Ks分布图呈现L分布, 分布图前段的峰值反映的是近期的基因复制.多倍化发生后会产生大量同源基因, 映射到Ks分布图上便会有大量Ks值接近的同源基因对产生.如果在分布图中有明显其他峰值存在, 则表明在这个Ks值的时间段内有大量的同源基因产生, 即全基因组复制事件(Lynch & Conery, 2000; Vanneste et al, 2013).另外, 假定同义替换以恒定速率堆积, 可以根据峰值对应的Ks值估算全基因组复制事件发生的时间(Badouin et al, 2017).根据公式T = Ks/2r可以推断物种的分化时间, 本研究r选用水蕨(Ceratopteris thalictroides)的1.104 × 10-8计算复制发生时间(Zhang et al, 2019). ...
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...
Regulation of translation initiation under abiotic stress conditions in plants: It is a conserved or not so conserved process among eukaryotes?
1
2012
... 通过对保留下来的复制基因进行功能富集分析(图2), 发现调控蛋白代谢过程(GO:0051246)、调控胞内蛋白代谢过程(GO:0032268)、调控翻译(GO:0006417)、基因表达转录后调控(GO:0010608)及翻译调控活性(GO:0045182)等GO terms被优势保留, 翼盖蕨可能通过调控转录、转录后调控、影响翻译起始因子活性, 并通过调控蛋白代谢, 参与逆境胁迫调控网络, 从而增强翼盖蕨对极端环境的适应性(Muñoz & Castellano, 2012; Guerra et al, 2015); 此外, 还富集到TATA-binding protein binding (GO:0017025), 其主要功能是通过控制Transcription factor IIB-related protein, 影响植物的萌发率(Niu et al, 2013).推测这些复制基因的偏倚性保留对于翼盖蕨的环境适应性起重要作用. ...
Pollen-expressed transcription factor 2 encodes a novel plant-specific TFIIB-related protein that is required for pollen germination and embryogenesis in Arabidopsis
1
2013
... 通过对保留下来的复制基因进行功能富集分析(图2), 发现调控蛋白代谢过程(GO:0051246)、调控胞内蛋白代谢过程(GO:0032268)、调控翻译(GO:0006417)、基因表达转录后调控(GO:0010608)及翻译调控活性(GO:0045182)等GO terms被优势保留, 翼盖蕨可能通过调控转录、转录后调控、影响翻译起始因子活性, 并通过调控蛋白代谢, 参与逆境胁迫调控网络, 从而增强翼盖蕨对极端环境的适应性(Muñoz & Castellano, 2012; Guerra et al, 2015); 此外, 还富集到TATA-binding protein binding (GO:0017025), 其主要功能是通过控制Transcription factor IIB-related protein, 影响植物的萌发率(Niu et al, 2013).推测这些复制基因的偏倚性保留对于翼盖蕨的环境适应性起重要作用. ...
One thousand plant transcriptomes and the phylogenomics of green plants
1
2019
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
A community-derived classification for extant lycophytes and ferns
2
2016
... 翼盖蕨科广布于泛热带地区, 目前被认为是单种科, 它是真水龙骨类I的基部类群(Zhang & Zhang, 2015; PPG I, 2016).鉴于翼盖蕨(Didymochlaena trancatula)特殊的系统位置, 本文通过对其进行转录组测序和Ks分析, 拟揭示: (1)翼盖蕨是否发生了全基因组复制事件? (2)什么时候发生的全基因组复制? (3)全基因组复制对翼盖蕨的物种形成及环境适应性的影响如何? ...
... 翼盖蕨的系统位置一直存在争议, 过去的研究认为翼盖蕨属于肿足蕨科成员(Smith & Cranfill, 2002; Schneider et al, 2004; Tsutsumi & Kato, 2005, 2006).近年来研究发现, 翼盖蕨属于真水龙骨类I (eupolypods I)的基部类群, 应该单独成立一科(Zhang & Zhang, 2015), 2016年PPG I接受翼盖蕨科的成立(PPG I, 2016). ...
The evolutionary history of ferns inferred from 25 low-copy nuclear genes
2
2015
... 本文通过对翼盖蕨进行全基因组复制分析, 发现翼盖蕨经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与翼盖蕨的物种分化时间基本一致(Rothfels et al, 2015); 此外, 翼盖蕨与同为真水龙骨类I的肿足蕨科、鳞毛蕨科等近缘类群的染色体基数一致, 均为X = 41 (Rothfels et al, 2015), 推测这次全基因组复制事件促进了翼盖蕨的物种分化及其物种多样性(Wang et al, 2012; Soltis et al, 2015). ...
... ); 此外, 翼盖蕨与同为真水龙骨类I的肿足蕨科、鳞毛蕨科等近缘类群的染色体基数一致, 均为X = 41 (Rothfels et al, 2015), 推测这次全基因组复制事件促进了翼盖蕨的物种分化及其物种多样性(Wang et al, 2012; Soltis et al, 2015). ...
Mining EST databases to resolve evolutionary events in major crop species
2
2004
... 本研究将Ks值小于0.1以及大于5的结果去除, 以排除随机误差以及同义替换饱和效应的影响(Blanc & Wolfe, 2004; Schlueter et al, 2004; Cui et al, 2006).为了避免峰的假阳性, 运用mclust中高斯混合模型对数据进行正态分布拟合(Fraley & Raftery, 2003). ...
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...
Ferns diversified in the shadow of angiosperms
1
2004
... 翼盖蕨的系统位置一直存在争议, 过去的研究认为翼盖蕨属于肿足蕨科成员(Smith & Cranfill, 2002; Schneider et al, 2004; Tsutsumi & Kato, 2005, 2006).近年来研究发现, 翼盖蕨属于真水龙骨类I (eupolypods I)的基部类群, 应该单独成立一科(Zhang & Zhang, 2015), 2016年PPG I接受翼盖蕨科的成立(PPG I, 2016). ...
Reciprocal gene loss between tetraodon and zebrafish after whole genome duplication in their ancestor
1
2007
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Intrafamilial relationships of the thelypteroid ferns (Thelypteridaceae)
1
2002
... 翼盖蕨的系统位置一直存在争议, 过去的研究认为翼盖蕨属于肿足蕨科成员(Smith & Cranfill, 2002; Schneider et al, 2004; Tsutsumi & Kato, 2005, 2006).近年来研究发现, 翼盖蕨属于真水龙骨类I (eupolypods I)的基部类群, 应该单独成立一科(Zhang & Zhang, 2015), 2016年PPG I接受翼盖蕨科的成立(PPG I, 2016). ...
Polyploidy and angiosperm diversification
1
2009
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Polyploidy and genome evolution in plants
1
2015
... 本文通过对翼盖蕨进行全基因组复制分析, 发现翼盖蕨经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与翼盖蕨的物种分化时间基本一致(Rothfels et al, 2015); 此外, 翼盖蕨与同为真水龙骨类I的肿足蕨科、鳞毛蕨科等近缘类群的染色体基数一致, 均为X = 41 (Rothfels et al, 2015), 推测这次全基因组复制事件促进了翼盖蕨的物种分化及其物种多样性(Wang et al, 2012; Soltis et al, 2015). ...
agriGO v2.0: A GO analysis toolkit for the agricultural community
1
2017
... 基于Ks分布结果, 调取峰值范围95%的基因, 对其进行功能注释(Ashburner et al, 2000).采用agriGO v2.0中的Fisher检验对保留下来的复制基因进行GO功能富集(中国农业大学, 北京) (P < 0.05) (Tian et al, 2017). ...
Molecular phylogenetic study on Davalliaceae
1
2005
... 翼盖蕨的系统位置一直存在争议, 过去的研究认为翼盖蕨属于肿足蕨科成员(Smith & Cranfill, 2002; Schneider et al, 2004; Tsutsumi & Kato, 2005, 2006).近年来研究发现, 翼盖蕨属于真水龙骨类I (eupolypods I)的基部类群, 应该单独成立一科(Zhang & Zhang, 2015), 2016年PPG I接受翼盖蕨科的成立(PPG I, 2016). ...
Evolution of epiphytes in Davalliaceae and related ferns
1
2006
... 翼盖蕨的系统位置一直存在争议, 过去的研究认为翼盖蕨属于肿足蕨科成员(Smith & Cranfill, 2002; Schneider et al, 2004; Tsutsumi & Kato, 2005, 2006).近年来研究发现, 翼盖蕨属于真水龙骨类I (eupolypods I)的基部类群, 应该单独成立一科(Zhang & Zhang, 2015), 2016年PPG I接受翼盖蕨科的成立(PPG I, 2016). ...
Indication of global deforestation at the Cretaceous-Tertiary boundary by New Zealand fern spike
1
2001
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...
The evolutionary significance of ancient genome duplications
1
2009
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary
1
2014
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...
Horsetails are ancient polyploids: Evidence from Equisetum giganteum
2
2015
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...
Inference of genome duplications from age distributions revisited
1
2013
... 目前全基因组复制事件检测常用的方法是同义替换率(Ks)法.它不影响氨基酸的正常编码, 为中性突变(Kimura, 1977), 假定其以近似恒定的速率累积, Ks值在某种程度上可指示同源基因的发生时间.正常情况下, 物种的Ks分布图呈现L分布, 分布图前段的峰值反映的是近期的基因复制.多倍化发生后会产生大量同源基因, 映射到Ks分布图上便会有大量Ks值接近的同源基因对产生.如果在分布图中有明显其他峰值存在, 则表明在这个Ks值的时间段内有大量的同源基因产生, 即全基因组复制事件(Lynch & Conery, 2000; Vanneste et al, 2013).另外, 假定同义替换以恒定速率堆积, 可以根据峰值对应的Ks值估算全基因组复制事件发生的时间(Badouin et al, 2017).根据公式T = Ks/2r可以推断物种的分化时间, 本研究r选用水蕨(Ceratopteris thalictroides)的1.104 × 10-8计算复制发生时间(Zhang et al, 2019). ...
Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio)
1
2012
... 本文通过对翼盖蕨进行全基因组复制分析, 发现翼盖蕨经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与翼盖蕨的物种分化时间基本一致(Rothfels et al, 2015); 此外, 翼盖蕨与同为真水龙骨类I的肿足蕨科、鳞毛蕨科等近缘类群的染色体基数一致, 均为X = 41 (Rothfels et al, 2015), 推测这次全基因组复制事件促进了翼盖蕨的物种分化及其物种多样性(Wang et al, 2012; Soltis et al, 2015). ...
Molecular evidence for an ancient duplication of the entire yeast genome
1
1997
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences,
1
2009
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication
1
2017
... 全基因组复制(whole genome duplication, WGD)或者多倍化(polyploid)是促进维管植物物种形成和适应性演化的重要动力之一(Van de Peer et al, 2009).植物界多数物种都经历了全基因组复制事件(Wolfe & Shields, 1997; Soltis et al, 2009; Huang et al, 2016; Xiang et al, 2017).研究显示, 25%的维管植物是多倍体(Barker et al, 2015), 其中15%的被子植物以及31%的蕨类植物以多倍体的形式存在(Wood et al, 2009; Jiao et al, 2011).现存种子植物和被子植物分化之前各经历了一次全基因组复制事件, 分别发生在3.19亿年前和1.92亿年前(Jiao et al, 2011), 即使基因组很小的拟南芥(Arabidopsis thaliana)也经历了多次全基因组复制事件(Athanasios et al, 2000; Bowers et al, 2003).全基因组复制使物种有了额外的基因组拷贝, 可以为物种进化提供更多的遗传原材料.全基因组复制发生以后, 复制基因将面临3个选择: 新功能化、亚功能化以及去功能化, 从而促进物种的多样性及其对极端环境的适应性(De Bodt et al, 2005; Semon & Wolfe, 2007; Hegarty & Hiscock, 2008; Freeling, 2009). ...
PAML 4: Phylogenetic analysis by maximum likelihood
1
2007
... 对物种内CDS序列进行all-against-all blast, 以e-5为阈值筛选基因家族中的旁系同源基因对, 使用mclblastline pipeline构建基因家族(Enright et al, 2002), 然后利用MUSCLE对每个基因家族进行比对(Edgar, 2004).运用PAML包中的CODEML软件计算得到Ks值(Goldman & Yang, 1994; Yang, 2007). ...
Didymochlaenaceae: A new fern family of eupolypods I (Polypodiales)
2
2015
... 翼盖蕨科广布于泛热带地区, 目前被认为是单种科, 它是真水龙骨类I的基部类群(Zhang & Zhang, 2015; PPG I, 2016).鉴于翼盖蕨(Didymochlaena trancatula)特殊的系统位置, 本文通过对其进行转录组测序和Ks分析, 拟揭示: (1)翼盖蕨是否发生了全基因组复制事件? (2)什么时候发生的全基因组复制? (3)全基因组复制对翼盖蕨的物种形成及环境适应性的影响如何? ...
... 翼盖蕨的系统位置一直存在争议, 过去的研究认为翼盖蕨属于肿足蕨科成员(Smith & Cranfill, 2002; Schneider et al, 2004; Tsutsumi & Kato, 2005, 2006).近年来研究发现, 翼盖蕨属于真水龙骨类I (eupolypods I)的基部类群, 应该单独成立一科(Zhang & Zhang, 2015), 2016年PPG I接受翼盖蕨科的成立(PPG I, 2016). ...
Dating whole genome duplication in Ceratopteris thalictroides and potential adaptive values of retained gene duplicates
3
2019
... 关于全基因组复制的研究目前主要集中在被子植物中, 而在陆生维管植物的重要代表——蕨类植物中鲜见报道.Dyer等(2013)基于细胞学观察间接推测了铁角蕨复合群(Asplenium monanthes complex)的多倍化情况.近年来, 随着分子生物学的快速发展, 基于Ks (synonymous substitution rates)或系统发育基因组学检测蕨类植物全基因组复制的报道在逐年增加, 如满江红(Azolla pinnata)(Li et al, 2018)、水蕨属(Ceratopteris spp.)(Barker, 2009; Zhang et al, 2019)、木贼属(Equisetum spp.)(Vanneste et al, 2015; Clark et al, 2019)以及基于大尺度系统发育框架的蕨类植物(Huang et al, 2019; One Thousand Plant Transcriptomes Initiative, 2019)的全基因组复制研究. ...
... 目前全基因组复制事件检测常用的方法是同义替换率(Ks)法.它不影响氨基酸的正常编码, 为中性突变(Kimura, 1977), 假定其以近似恒定的速率累积, Ks值在某种程度上可指示同源基因的发生时间.正常情况下, 物种的Ks分布图呈现L分布, 分布图前段的峰值反映的是近期的基因复制.多倍化发生后会产生大量同源基因, 映射到Ks分布图上便会有大量Ks值接近的同源基因对产生.如果在分布图中有明显其他峰值存在, 则表明在这个Ks值的时间段内有大量的同源基因产生, 即全基因组复制事件(Lynch & Conery, 2000; Vanneste et al, 2013).另外, 假定同义替换以恒定速率堆积, 可以根据峰值对应的Ks值估算全基因组复制事件发生的时间(Badouin et al, 2017).根据公式T = Ks/2r可以推断物种的分化时间, 本研究r选用水蕨(Ceratopteris thalictroides)的1.104 × 10-8计算复制发生时间(Zhang et al, 2019). ...
... 研究发现, 许多物种的全基因组复制事件聚集在白垩纪的物种灭绝时期(60-70 Mya)(Lynch & Conery, 2000; Schlueter et al, 2004; Cui et al, 2006), 该时期地球环境发生剧烈变化, 物种大规模灭绝, 部分物种能适应这种极端环境并存活至今可能归因于物种的全基因组加倍(Vajda et al, 2001; Fawcett et al, 2009).本研究发现翼盖蕨至少经历了两次全基因组复制事件(图3), 其中一次发生在90-94 Mya, 这与已报道的巨木贼(Equisetum giganteum)中全基因组复制发生时间相一致(Vanneste et al, 2015); 另一次发生于59-62 Mya (Cretaceous-Tertiary C-T大灭绝事件), 这与水蕨中报道的全基因组复制发生时间一致, 推测翼盖蕨的这次全基因组复制事件使其成功度过物种大灭绝时期并存活下来(Vanneste et al, 2014; Zhang et al, 2019).然而, 全基因组复制如何促进翼盖蕨对环境的适应性? ...