

高通量测序技术在线虫多样性研究中的应用
收稿日期: 2022-05-13
录用日期: 2022-07-14
网络出版日期: 2022-09-09
基金资助
中国科学院战略性先导科技专项子课题(XDA28020202);国家自然科学基金(41877047);中国科学院国际合作重点项目(151221KYSB20200014)
Application of high-throughput sequencing technique in the study of nematode diversity
Received date: 2022-05-13
Accepted date: 2022-07-14
Online published: 2022-09-09
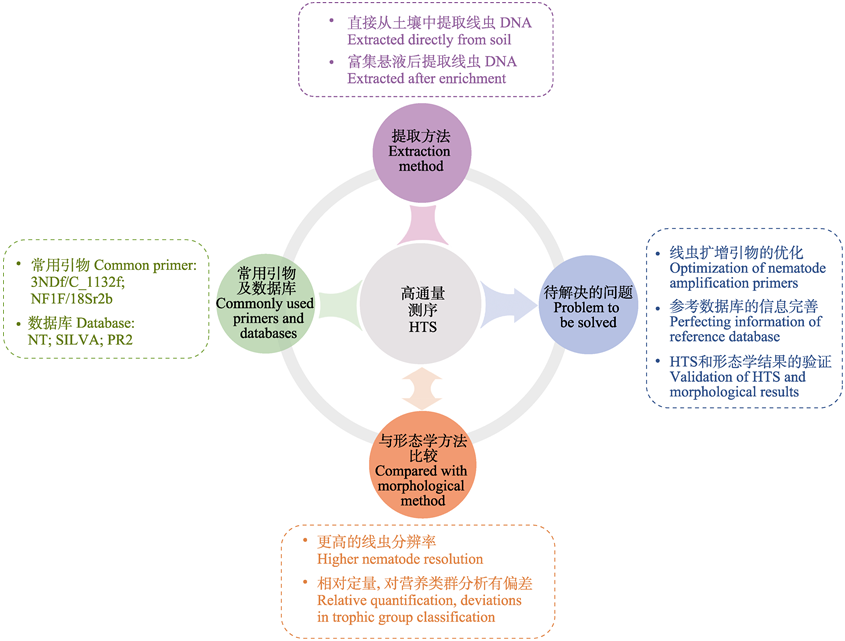
土壤线虫多样性是土壤生态学研究的热点之一, 然而对土壤线虫群落组成及多样性的研究通常受到分类学和方法学的限制。当前, 分子生物学技术的快速发展丰富了我们对土壤线虫多样性的认识, 但也存在一定的局限性。本文综述了常用分子生物学技术如变性梯度凝胶电泳(denaturing gradient gel electrophoresis, DGGE)、末端限制性片段长度多态性分析(terminal restriction fragment length polymorphism, T-RFLP)、实时荧光定量PCR (quantitative real-time PCR, qPCR)和高通量测序(high-throughput sequencing, HTS)技术近年来在线虫多样性研究中的应用, 重点从土壤线虫DNA提取方法、引物和数据库的选择、高通量测序技术和形态学鉴定结果的比较等方面阐述了高通量测序技术在线虫多样性研究中的优势与不足, 并提出选择合适的线虫DNA提取方法结合特定引物和数据库进行注释分析, 仍是今后使用高通量测序技术开展线虫多样性研究的重点。当研究目标是土壤线虫多样性时, 优先推荐富集线虫悬液提取DNA的方法, 因此, 研究人员应根据具体目标选择最优组合开展实验研究。
孙翌昕, 李英滨, 李玉辉, 李冰, 杜晓芳, 李琪 . 高通量测序技术在线虫多样性研究中的应用[J]. 生物多样性, 2022 , 30(12) : 22266 . DOI: 10.17520/biods.2022266
Background & Aims: Soil nematode diversity is one of the hot topic in soil ecology research. However, in-depth studies on soil nematode community composition and diversity are limited by the established taxonomy and methodology. At present, the development of molecular biological techniques has elevated our understanding of the diversity of soil nematodes, but it also included set-backs. This paper summarizes the research progress in molecular biology technology, including denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphism (T-RFLP), quantitative real-time PCR (qPCR) and high-throughput sequencing (HTS) technology, and the advantages and disadvantages of high-throughput sequencing in nematode research. We elaborate these techniques based on the extraction of soil nematode DNA, the selection of primers and reference databases, as well as the comparison between high-throughput sequencing and morphological identification analysis.
Prospects: The selections of soil nematode DNA extraction methods, in combination with proper primers and reference databases remain the key point to study nematode diversity. When focusing on nematode classification, we recommend DNA extraction after enrichment of nematodes suspension. Conclusively, researchers can then select the optimal combination to conduct experiments according to their research objectives.

| [1] | Abad D, Albaina A, Aguirre M, Laza-Martínez A, Uriarte I, Iriarte A, Villate F, Estonba A (2016) Is metabarcoding suitable for estuarine plankton monitoring? A comparative study with microscopy. Marine Biology, 163, 1-13. |
| [2] | Ahmed M, Back MA, Prior T, Karssen G, Lawson R, Adams I, Sapp M (2019) Metabarcoding of soil nematodes: The importance of taxonomic coverage and availability of reference sequences in choosing suitable marker(s). Metabarcoding and Metagenomics, 3, 77-99. |
| [3] | Bhadury P, Austen MC, Bilton DT, Lambshead PJD, Rogers AD, Smerdon GR (2005) Combined morphological and molecular analysis of individual nematodes through short-term preservation in formalin. Molecular Ecology Notes, 5, 965-968. |
| [4] | Chen XY, Daniell TJ, Neilson R, O’Flaherty V, Griffiths BS (2010) A comparison of molecular methods for monitoring soil nematodes and their use as biological indicators. European Journal of Soil Biology, 46, 319-324. |
| [5] | Cook AA, Bhadury P, Debenham NJ, Meldal BHM, Blaxter ML, Smerdon GR, Austen MC, Lambshead PJD, Rogers AD (2005) Denaturing gradient gel electrophoresis (DGGE) as a tool for identification of marine nematodes. Marine Ecology Progress Series, 291, 103-113. |
| [6] | Creer S, Fonseca VG, Porazinska DL, Giblin-Davis RM, Sung W, Power DM, Packer M, Carvalho GR, Blaxter ML, Lambshead PJD, Thomas WK (2010) Ultrasequencing of the meiofaunal biosphere: Practice, pitfalls and promises. Molecular Ecology, 19, 4-20. |
| [7] | Donn S, Neilson R, Griffiths BS, Daniell TJ (2012) A novel molecular approach for rapid assessment of soil nematode assemblages—Variation, validation and potential applications. Methods in Ecology and Evolution, 3, 12-23. |
| [8] | Drummond AJ, Newcomb RD, Buckley TR, Xie D, Dopheide A, Potter BCM, Heled J, Ross HA, Tooman L, Grosser S, Park D, Demetras NJ, Stevens MI, Russell JC, Anderson SH, Carter A, Nelson N (2015) Evaluating a multigene environmental DNA approach for biodiversity assessment. GigaScience, 4, 46. |
| [9] | Du XF, Li YB, Han X, Ahmad W, Li Q (2020) Using high-throughput sequencing quantitatively to investigate soil nematode community composition in a steppe-forest ecotone. Applied Soil Ecology, 152, 103562. |
| [10] | Du XF, Li YB, Liu F, Su XL, Li Q (2018) Structure and ecological functions of soil micro-food web. Chinese Journal of Applied Ecology, 29, 403-411. (in Chinese with English abstract) |
| [10] | [ 杜晓芳, 李英滨, 刘芳, 宿晓琳, 李琪 (2018) 土壤微食物网结构与生态功能. 应用生态学报, 29, 403-411.] |
| [11] | Du XF, Liang WJ, Li Q (2021) DNA extraction, amplification and high-throughput sequencing of soil nematode community. Microbiome Protocols eBook, Bio-101, e2104085. (in Chinese) |
| [11] | [ 杜晓芳, 梁文举, 李琪 (2021) 土壤线虫群落DNA提取、扩增及高通量测序. 微生物组实验手册, Bio-101, e2104085.] |
| [12] | Du XF, Liu HW, Li YB, Li B, Han X, Li YH, Mahamood M, Li Q (2022) Soil community richness and composition jointly influence the multifunctionality of soil along the forest-steppe ecotone. Ecological Indicators, 139, 108900. |
| [13] | Edel-Hermann V, Gautheron N, Alabouvette C, Steinberg C (2008) Fingerprinting methods to approach multitrophic interactions among microflora and microfauna communities in soil. Biology and Fertility of Soils, 44, 975-984. |
| [14] | Foucher A, Bongers T, Noble LR, Wilson M (2004) Assessment of nematode biodiversity using DGGE of 18S rDNA following extraction of nematodes from soil. Soil Biology and Biochemistry, 36, 2027-2032. |
| [15] | Foucher A, Wilson M (2002) Development of a polymerase chain reaction-based denaturing gradient gel electrophoresis technique to study nematode species biodiversity using the 18s rDNA gene. Molecular Ecology Notes, 2, 45-48. |
| [16] | Gao DD, Moreira-Grez B, Wang KL, Zhang W, Xiao SS, Wang WL, Chen HS, Zhao J (2021) Effects of ecosystem disturbance on nematode communities in calcareous and red soils: Comparison of taxonomic methods. Soil Biology and Biochemistry, 155, 108162. |
| [17] | Geisen S, Snoek LB ten Hooven FC, Duyts H, Kostenko O, Bloem J, Martens H, Quist CW, Helder JA,van der Putten WH (2018) Integrating quantitative morphological and qualitative molecular methods to analyse soil nematode community responses to plant range expansion. Methods in Ecology and Evolution, 9, 1366-1378. |
| [18] | George PBL, Lindo Z (2015) Congruence of community structure between taxonomic identification and T-RFLP analyses in free-living soil nematodes. Pedobiologia, 58, 113-117. |
| [19] | Griffiths BS, de Groot GA, Laros I, Stone D, Geisen S (2018) The need for standardisation: Exemplified by a description of the diversity, community structure and ecological indices of soil nematodes. Ecological Indicators, 87, 43-46. |
| [20] | Griffiths BS, Donn S, Neilson R, Daniell TJ (2006) Molecular sequencing and morphological analysis of a nematode community. Applied Soil Ecology, 32, 325-337. |
| [21] | Holeva R, Phillips MS, Neilson R, Brown DJF, Young V, Boutsika K, Blok VC (2006) Real-time PCR detection and quantification of vector trichodorid nematodes and tobacco rattle virus. Molecular and Cellular Probes, 20, 203-211. |
| [22] | Hou L, Xue B, Xue HY (2019) Study on soil nematode community of Northern Tibetan alpine meadow under simulated warming condition by high-throughput sequencing method. Acta Agrestia Sinica, 27, 443-451. (in Chinese with English abstract) |
| [22] | [ 侯磊, 薛蓓, 薛会英 (2019) 利用高通量测序法对模拟增温条件下藏北高寒草甸土壤线虫群落的研究. 草地学报, 27, 443-451.] |
| [23] | Kawanobe M, Toyota K, Ritz K (2021) Development and application of a DNA metabarcoding method for comprehensive analysis of soil nematode communities. Applied Soil Ecology, 166, 103974. |
| [24] | Kenmotsu H, Takabayashi E, Takase A, Hirose Y, Eki T(2021) Use of universal primers for the 18S ribosomal RNA gene and whole soil DNAs to reveal the taxonomic structures of soil nematodes by high-throughput amplicon sequencing. PLoS ONE, 16, e0259842. |
| [25] | Kitagami Y, Obase K, Matsuda Y (2022) High-throughput sequencing and conventional morphotyping show different soil nematode assemblages but similar community responses to altitudinal gradients on Mt. Ibuki, Japan. Pedobiologia, 90, 150788. |
| [26] | Kushida A (2013) Design and evaluation of PCR primers for denaturing gradient gel electrophoresis analysis of plant parasitic and fungivorous nematode communities. Microbes and Environments, 28, 269-274. |
| [27] | Li B, Li YB, Fanin N, Han X, Du XF, Liu HW, Li YH, Li Q (2022) Adaptation of soil micro-food web to elemental limitation: Evidence from the forest-steppe ecotone. Soil Biology and Biochemistry, 170, 108698. |
| [28] | Li Q, Liang WJ, Zhang XK, Mahamood M (2017) Soil Nematodes of Grasslands in Northern China. Zhejiang University Press, Hangzhou & Elsevier Academic Press, Amsterdam. |
| [29] | Li YB, Liang SW, Du XF, Kou XC, Lv XT, Li Q (2021) Mowing did not mitigate the negative effects of nitrogen deposition on soil nematode community in a temperate steppe. Soil Ecology Letters, 3, 125-133. |
| [30] | Liu HW, Du XF, Li YB, Han X, Li B, Zhang XK, Li Q, Liang WJ (2022) Organic substitutions improve soil quality and maize yield through increasing soil microbial diversity. Journal of Cleaner Production, 347, 131323. |
| [31] | Lott MJ, Hose GC, Power ML (2014) Towards the molecular characterisation of parasitic nematode assemblages: An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis. Experimental Parasitology, 144, 76-83. |
| [32] | Madani M, Subbotin SA, Moens M (2005) Quantitative detection of the potato cyst nematode, Globodera pallida, and the beet cyst nematode, Heterodera schachtii, using real-time PCR with SYBR green I dye. Molecular and Cellular Probes, 19, 81-86. |
| [33] | Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction- amplified genes coding for 16S rRNA. Applied and Environmental Microbiology, 59, 695-700. |
| [34] | Nakhla MK, Owens KJ, Li WB, Wei G, Skantar AM, Levy L (2010) Multiplex real-time PCR assays for the identification of the potato cyst and tobacco cyst nematodes. Plant Disease, 94, 959-965. |
| [35] | Nisa RU, Tantray AY, Shah AA (2022) Shift from morphological to recent advanced molecular approaches for the identification of nematodes. Genomics, 114, 110295. |
| [36] | Ovre?s L, Forney L, Daae FL, Torsvik V (1997) Distribution of bacterioplankton in meromictic lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Applied and Environmental Microbiology, 63, 3367-3373. |
| [37] | Peham T, Steiner FM, Schlick-Steiner BC, Arthofer W (2017) Are we ready to detect nematode diversity by next generation sequencing? Ecology and Evolution, 7, 4147-4151. |
| [38] | Powers T (2004) Nematode molecular diagnostics: From bands to barcodes. Annual Review of Phytopathology, 42, 367-383. |
| [39] | Rettedal EA, Clay S, Br?zel VS (2010) GC-clamp primer batches yield 16S rRNA gene amplicon pools with variable GC clamps, affecting denaturing gradient gel electrophoresis profiles. FEMS Microbiology Letters, 312, 55-62. |
| [40] | Sapkota R, Nicolaisen M (2015) High-throughput sequencing of nematode communities from total soil DNA extractions. BMC Ecology, 15, 3. |
| [41] | Schenk J, Geisen S, Kleinb?lting N, Traunspurger W (2019) Metabarcoding data allow for reliable biomass estimates in the most abundant animals on earth. Metabarcoding and Metagenomics, 3, e46704. |
| [42] | Schenk J, Kleinb?lting N, Traunspurger W (2020) Comparison of morphological, DNA barcoding, and metabarcoding characterizations of freshwater nematode communities. Ecology and Evolution, 10, 2885-2899. |
| [43] | Sikder MM, Vesterg?rd M, Sapkota R, Kyndt T, Nicolaisen M (2020) Evaluation of metabarcoding primers for analysis of soil nematode communities. Diversity, 12, 388. |
| [44] | Sun YX, Du XF, Li YB, Han X, Fang S, Geisen S, Li Q (2023) Database and primer selections affect nematode community composition under different vegetations of Changbai Mountain. Soil Ecology Letters, 5, 142-150. |
| [45] | Tedersoo L, Drenkhan R, Anslan S, Morales-Rodriguez C, Cleary M (2019) High-throughput identification and diagnostics of pathogens and pests: Overview and practical recommendations. Molecular Ecology Resources, 19, 47-76. |
| [46] | Treonis AM, Unangst SK, Kepler RM, Buyer JS, Cavigelli MA, Mirsky SB, Maul JE (2018) Characterization of soil nematode communities in three cropping systems through morphological and DNA metabarcoding approaches. Scientific Reports, 8, 2004. |
| [47] | Waeyenberge L, de Sutter N, Viaene N, Haegeman A (2019) New insights into nematode DNA-metabarcoding as revealed by the characterization of artificial and spiked nematode communities. Diversity, 11, 1-22. |
| [48] | Wang IC (2012) Developing a qPCR-based Molecular Technique for Nematode Community Analysis. PhD dissertation, University of Hawaii, Hawaii. |
| [49] | Wang N, Huang JH, Huo N, Yang PP, Zhang XY, Zhao SW (2021) Characteristics of soil nematode community under different vegetation restoration approaches in the mountainous region of southern Ningxia: A comparative study based on morphological identification and high-throughput sequencing methods. Biodiversity Science, 29, 1513-1529. (in Chinese with English abstract) |
| [49] | [ 王楠, 黄菁华, 霍娜, 杨盼盼, 张欣玥, 赵世伟 (2021) 宁南山区不同植被恢复方式下土壤线虫群落特征: 形态学鉴定与高通量测序法比较. 生物多样性, 29, 1513-1529.] |
| [50] | Wang SB, Li Q, Liang WJ, Jiang Y, Jiang SW (2008) PCR- DGGE analysis of nematode diversity in Cu-contaminated soil. Pedosphere, 18, 621-627. |
| [51] | Wiesel L, Daniell TJ, King D, Neilson R (2015) Determination of the optimal soil sample size to accurately characterise nematode communities in soil. Soil Biology and Biochemistry, 80, 89-91. |
/
| 〈 |
|
〉 |