三种蚜虫的线粒体基因组数据
- 闽台作物有害生物生态防控国家重点实验室, 福建农林大学植物保护学院, 福州 350002
收稿日期: 2022-04-20
录用日期: 2022-06-13
网络出版日期: 2022-06-24
基金资助
国家自然科学基金(31970446)
Mitochondrial genome data of three aphid species
- State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou 350002
Received date: 2022-04-20
Accepted date: 2022-06-13
Online published: 2022-06-24
本文引用格式
卢聪聪 , 刘倩 , 黄晓磊 . 三种蚜虫的线粒体基因组数据[J]. 生物多样性, 2022 , 30(7) : 22204 . DOI: 10.17520/biods.2022204
Abstract
The complete mitochondrial genomes (mitogenomes) have been widely used in the studies of molecular evolution, genomics and phylogeny. Aphids are important agricultural and forestry pests. Considering the reported complete mitogenomes of aphids are still limited, it will be of great value to obtain more aphid mitogenome data for related researches. This paper reports the complete mitogenome sequences of three aphid species, Greenidea ficicola, Aphis aurantii and Mindarus keteleerifoliae. The data of their detailed annotation, their gene sequence and gene structure, and the codon usage are also presented. These data can benefit future researches such as insect phylogenetic relationships, population divergence patterns as well as insect pest control.
Database/Dataset Profile
| Title | Mitochondrial genome data of three aphid species |
|---|---|
| Authors | Congcong Lu, Qian Liu, Xiaolei Huang |
| Corresponding author | Xiaolei Huang (huangxl@fafu.edu.cn) |
| Time range | 2015-2017 |
| Geographical scope | Quanzhou (Fujian), Fuzhou (Fujian) |
| File size | 48 MB |
| Data format | *.fasta, *.xlsx, *.tif |
| Data link | |
| Database/Dataset composition | The dataset consists of 7 data files in total, containing 3 sequence files, with a data volume of 50 KB; one excel data files, with a data volume of 37 KB; and 3 picture files, with a data volume of 47.4 MB. |
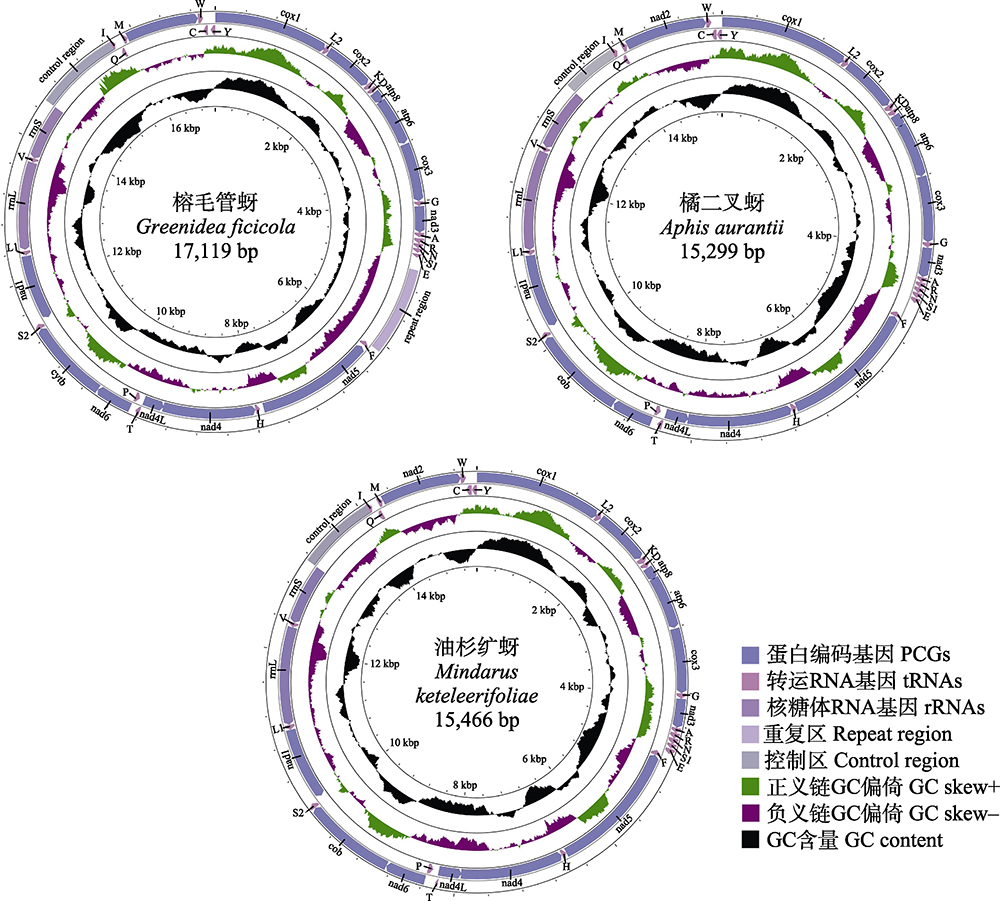
Key words: aphids; mitochondrial genome; gene annotation; genomic data
参考文献
| [1] | Al Arab M, Zu Siederdissen CH, Tout K, Sahyoun AH, Stadler PF, Bernt M (2017) Accurate annotation of protein-coding genes in mitochondrial genomes. Molecular Phylogenetics and Evolution, 106, 209-216. |
| [2] | Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF (2013) MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69, 313-319. |
| [3] | Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Research, 27, 1767-1780. |
| [4] | Burger G, Gray MW, Lang BF (2003) Mitochondrial genomes: Anything goes. Trends in Genetics, 19, 709-716. |
| [5] | Chen J, Wang Y, Jiang LY, Qiao GX (2017) Mitochondrial genome sequences effectively reveal deep branching events in aphids (Insecta: Hemiptera: Aphididae). Zoologica Scripta, 46, 706-717. |
| [6] | Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45, e18. |
| [7] | Favret C (2022) Aphid species file, Version 5.0/5.0. http://Aphid.SpeciesFile.org. (accessed on 2022-01-24) |
| [8] | Grant JR, Stothard P (2008) The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Research, 36, W181-W184. |
| [9] | Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874. |
| [10] | Laslett D, Canbäck B (2008) ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics, 24, 172-175. |
| [11] | Li H, Leavengood Jr JM, Chapman EG, Burkhardt D, Song F, Jiang P, Liu JP, Zhou XG, Cai WZ (2017) Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proceedings of the Royal Society B: Biological Sciences, 284, 20171223. |
| [12] | Liu YQ, Li H, Song F, Zhao YS, Wilson JJ, Cai WZ (2019) Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Systematic Entomology, 44, 810-819. |
| [13] | Lorenz R, Bernhart SH, Zu Siederdissen CH, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) ViennaRNA Package 2.0. Algorithms for Molecular Biology, 6, 26. |
| [14] | Ma C, Yang P, Jiang F, Chapuis MP, Shali Y, Sword GA, Kang L (2012) Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Molecular Ecology, 21, 4344-4358. |
| [15] | Mueller RL (2006) Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Systematic Biology, 55, 289-300. |
| [16] | Nass MMK, Nass S (1963) Intramitochondrial fibers with DNA characteristics: I. Fixation and electron staining reactions. The Journal of Cell Biology, 19, 593-611. |
| [17] | Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng QD, Wortman J, Young SK, Earl AM (2014) Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE, 9, e112963. |
| [18] | Wang Y, Huang XL, Qiao GX (2014) The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae). Insect Science, 21, 278-290. |
| [19] | Wilson AC, Cann RL, Carr SM, George M, Gyllensten UB, Helm-Bychowski KM, Higuchi RG, Palumbi SR, Prager EM, Sage RD, Stoneking M (1985) Mitochondrial DNA and two perspectives on evolutionary genetics. Biological Journal of the Linnean Society, 26, 375-400. |
| [20] | Wolstenholme DR (1992) Animal mitochondrial DNA: Structure and evolution. International Review of Cytology, 141, 173-216. |
| [21] | Zhang B, Ma C, Edwards O, Fuller S, Kang L (2014) The mitochondrial genome of the Russian wheat aphid Diuraphis noxia: Large repetitive sequences between trnE and trnF in aphids. Gene, 533, 253-260. |
| [22] | Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348-355. |
| [23] | Zhang H, Liu Q, Lu CC, Deng J, Huang XL (2021) The first complete mitochondrial genome of Lachninae species and comparative genomics provide new insights into the evolution of gene rearrangement and the repeat region. Insects, 12, 55. |