七种半翅目昆虫线粒体基因组数据
- 1.农林生物安全全国重点实验室, 福建农林大学植物保护学院, 福州 350002
2.中国科学院动物研究所动物进化与系统学重点实验室, 北京 100101
收稿日期: 2024-10-01
录用日期: 2024-12-10
网络出版日期: 2025-03-14
基金资助
福建农林大学科技创新专项基金(KFB23016);福建农林大学杰出青年科研人才计划项目(Kxjq21004)
Mitochondrial genomic data for seven Hemipteran species
- Lin Zhen ,
- Xiang Jiabao ,
- Cai Hejiayi ,
- Gao Bei ,
- Yang Jintao ,
- Li Junyi ,
- Zhou Qingsong ,
- Huang Xiaolei ,
- Deng Jun
- 1 State Key Laboratory of Agricultural and Forestry Biosecurity, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou 350002, China
2 Key Laboratory of Zoological Systematics and Evolution, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China
Received date: 2024-10-01
Accepted date: 2024-12-10
Online published: 2025-03-14
Supported by
Science and Technology Innovation Special Fund of Fujian Agriculture and Forestry University(KFB23016);Outstanding Young Scientific Research Talents Program of Fujian Agriculture and Forestry University(Kxjq21004)
摘要
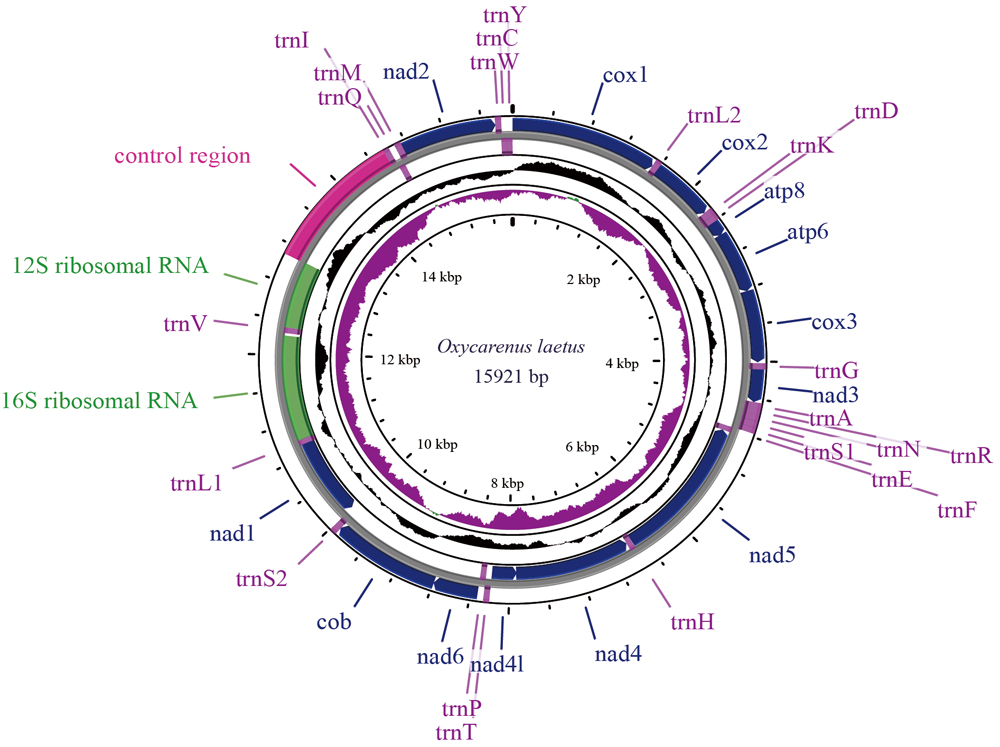
昆虫是地球上物种多样性最高的类群, 对特定昆虫类群的线粒体基因组研究还处于不断发展和积累的阶段。由于部分半翅目昆虫的线粒体基因组在GenBank数据库中缺失, 阻碍了线粒体基因组在半翅目昆虫中的进一步应用。本研究报道了Maevius indecorus、Limnogonus cereiventris、尖长蝽(Oxycarenus laetus)、Hormaphis cornu、Melanaphis donacis、香蕉交脉蚜(Pentalonia nigronervosa)、石榴囊毡蚧(Acanthococcus lagerstroemiae)等7种半翅目昆虫的线粒体基因组详尽信息, 其中Maevius indecorus和Pentalonia nigronervosa线粒体基因组数据分别为锚蝽科和交脉蚜属(Pentalonia)首次报道。7种半翅目昆虫数据通过SRA数据库获得, 来源于6个国家(澳大利亚、喀麦隆、中国、印度、肯尼亚、美国)的7个地区。本研究可为半翅目昆虫系统发育关系、生物多样性、种群分化格局等相关研究提供一定数据支持。
数据库(集)基本信息简介
| 数据库(集)名称 | 七种半翅目昆虫线粒体基因组数据 |
|---|---|
| 作者 | 林珍, 向家宝, 蔡何佳奕, 高贝, 杨金涛, 李俊毅, 周青松, 黄晓磊, 邓鋆 |
| 通讯作者 | 黄晓磊(huangxl@fafu.edu.cn); 邓鋆(tiancai1282008@126.com) |
| 时间范围 | 2019-2024年 |
| 地理区域 | 澳大利亚、喀麦隆、中国、印度、肯尼亚、美国 |
| 文件大小 | 1.8 MB |
| 数据格式 | *.fasta, *.xlsx, *.png |
| 数据链接 | https://doi.org/10.57760/sciencedb.21938 https://www.biodiversity-science.net/fileup/1005-0094/DATA/2024434.zip |
| 数据库(集)组成 | 数据集共由15个文件组成, 包含: 7个序列数据文件, 文件大小108 KB; 1个excel数据文件, 文件大小75 KB; 7个图片数据文件, 文件大小1.62 MB。 |
本文引用格式
林珍 , 向家宝 , 蔡何佳奕 , 高贝 , 杨金涛 , 李俊毅 , 周青松 , 黄晓磊 , 邓鋆 . 七种半翅目昆虫线粒体基因组数据[J]. 生物多样性, 2025 , 33(2) : 24434 . DOI: 10.17520/biods.2024434
Abstract
Insects represent the most diverse group of organisms on Earth, and research on the mitochondrial genomes of specific insect groups remains in the stages of development and accumulation. A notable lack of mitochondrial genome data for certain Hemiptera species in the GenBank database has hindered further exploration and applications in this field. This study reports detailed mitochondrial genome information for seven Hemipteran species: Maevius indecorus, Limnogonus cereiventris, Oxycarenus laetus, Hormaphis cornu, Melanaphis donacis, Pentalonia nigronervosa, and Acanthococcus lagerstroemiae. Among these species, Maevius indecorus and Pentalonia nigronervosa represent the first complete mitochondrial genomes reported for the family Hyocephalidae and the genus Pentalonia, respectively. The mitochondrial genome data for these seven species were retrieved from the SRA database, encompassing seven regions across six countries: Australia, Cameroon, China, India, Kenya, and the United States. This dataset provides valuable support for research on phylogenetic relationships, biodiversity, and population differentiation within certain Hemiptera species.
Database/Dataset Profile
| Title | Mitochondrial genomic data for seven Hemipteran species |
|---|---|
| Data author(s) | Zhen Lin, Jiabao Xiang, Hejiayi Cai, Bei Gao, Jintao Yang, Junyi Li, Qingsong Zhou, Xiaolei Huang, Jun Deng |
| Data corresponding author | Xiaolei Huang (huangxl@fafu.edu.cn); Jun Deng (tiancai1282008@126.com) |
| Time range | 2019-2024 |
| Geographical scope | Australia, Cameroon, China, India, Kenya, USA |
| File size | 1.8 MB |
| Data format | *.fasta, *.xlsx, *.png |
| Data link | https://doi.org/10.57760/sciencedb.21938 https://www.biodiversity-science.net/fileup/1005-0094/DATA/2024434.zip |
| Database/Dataset composition | The dataset consists of 15 data files in total, containing 7 sequence files with a combined size of 108 KB, 7 figure files with a combined size of 1.62 MB, and one excel data file with a size of 75 KB. |

Key words: Hemiptera; mitochondrial genome; gene annotation; biodiversity
参考文献
| [1] | Bánki O, Roskov Y, D?ring M, Ower G, Hernández Robles DR, Plata Corredor CA, Stjernegaard Jeppesen T, ?rn A, Pape T, Hobern D, Garnett S, Little H, DeWalt RE, Ma K, Miller J, Orrell T, Aalbu R, Abbott J, Aedo C, et al. (2024) Catalogue of Life (Version 2024-08-29). Catalogue of Life, Amsterdam, Netherlands. https://doi.org/10.48580/dgdwl. |
| [2] | Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF (2013) MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69, 313-319. |
| [3] | Burger G, Gray MW, Franz Lang B (2003) Mitochondrial genomes: Anything goes. Trends in Genetics, 19, 709-716. |
| [4] | Dierckxsens N, Mardulyn P, Smits G (2017) NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45, e18. |
| [5] | Grant JR, Stothard P (2008) The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Research, 36, W181-W184. |
| [6] | Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870-1874. |
| [7] | Li DH, Liu CM, Luo RB, Sadakane K, Lam TW (2015) MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics, 31, 1674-1676. |
| [8] | Lu CC, Huang XL, Deng J (2020) The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. International Journal of Biological Macromolecules, 158, 854-864. |
| [9] | Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng QD, Wortman J, Young SK, Earl AM (2014) Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE, 9, e112963. |
| [10] | Zhang D, Gao FL, Jakovli? I, Zou H, Zhang J, Li WX, Wang GT (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20, 348-355. |